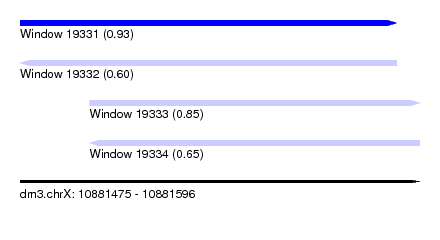
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,881,475 – 10,881,596 |
| Length | 121 |
| Max. P | 0.930480 |

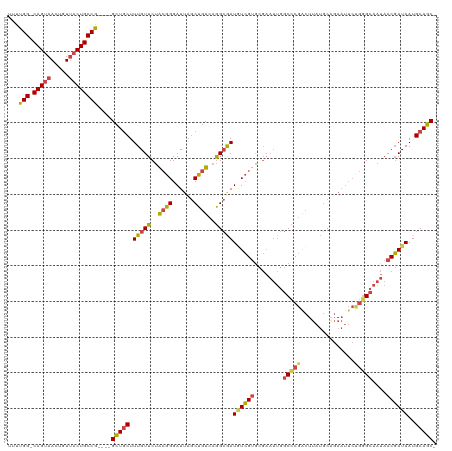
| Location | 10,881,475 – 10,881,589 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Shannon entropy | 0.31967 |
| G+C content | 0.48254 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10881475 114 + 22422827 -GCUGCAUUAUCGUUAUUGCCCAGAAUUCUGCAUAAGUCGGGCCAAUUCGCACGACAUUGCCUCACAGCUAAUCCCGCAGAACAAAGCAGC----CGGCAGUAACAGUUACUG-CCAAAA -(((((......((....))......((((((....((((.((......)).))))...((......)).......))))))....)))))----.(((((((.....)))))-)).... ( -35.20, z-score = -3.07, R) >droAna3.scaffold_13417 5902499 102 - 6960332 GGUUGCAUUAUAGUUAUAGC-------UUCGUGAGAGUGGAG-------GUCCUAGAUC-CUUCAGAGU--AUCCU-UAGAACAAAGUAACAGUUUGGCAGGCAAAGUUUCUGGCCAAAA .(((((..............-------(((....)))(((((-------(........)-)))))....--.....-.........)))))..(((((((((.......))).)))))). ( -22.40, z-score = 0.21, R) >droEre2.scaffold_4690 14501993 114 - 18748788 -GCUGCAUUAUCGUUAUUGCCCGGAAUUCUGCAUAAGCCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGCAGAACAAAGCAGC----CGGCAGUAACAGUUACUG-CCAAAA -(((((....(((........)))..((((((((.((((((((....((....))....)))))...))).))...))))))....)))))----.(((((((.....)))))-)).... ( -35.30, z-score = -2.09, R) >droYak2.chrX 10528046 114 - 21770863 -GCUGCAUUAUCGUUAUUGCCCAGAAUUCAGCAUAAGUCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGUGGAACAAAGCAGC----CGGCAGUAACAGUUACUG-CCAAAA -(((((......(((..(((..........)))...(((((((....((....))....))))))))))...(((...))).....)))))----.(((((((.....)))))-)).... ( -34.10, z-score = -2.28, R) >droSec1.super_15 1583473 114 + 1954846 -GCUGCAUUAUCGUUAUUGCCCAGAAUUCUGCAUAAGUCGAGCCAAUUCGCACGAUAUCGCCUGACAGCUAAUCCCGUAGAACAAAGCAGC----CGGCAGUAACAGUUACUG-CCAAAA -(((((......((....))......((((((....((((.((......)).))))...((......)).......))))))....)))))----.(((((((.....)))))-)).... ( -31.30, z-score = -2.62, R) >droSim1.chrX 8434774 114 + 17042790 -GCUGCAUUAUCGUUAUUGCCCAGAAUUCUGCAUAAGUCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGUAGAACAAAGCAGC----CGGCAGUAACAGUUACUG-CCAAAA -(((((......((....))......((((((....(((((((....((....))....))))))).(.......)))))))....)))))----.(((((((.....)))))-)).... ( -34.80, z-score = -2.85, R) >consensus _GCUGCAUUAUCGUUAUUGCCCAGAAUUCUGCAUAAGUCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGUAGAACAAAGCAGC____CGGCAGUAACAGUUACUG_CCAAAA .(((((......((....))......((((((....((((.((......)).))))...((......)).......))))))....))))).....(((((((.....))))).)).... (-22.39 = -22.32 + -0.08)

| Location | 10,881,475 – 10,881,589 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.99 |
| Shannon entropy | 0.31967 |
| G+C content | 0.48254 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.21 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
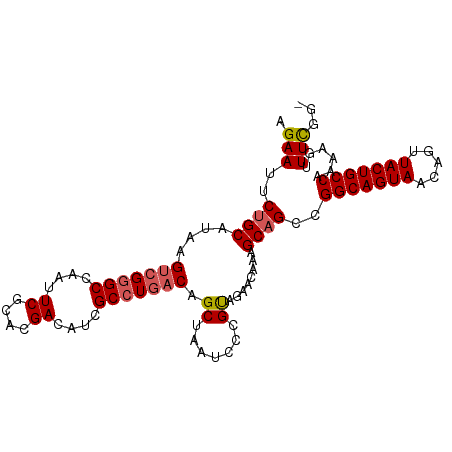
>dm3.chrX 10881475 114 - 22422827 UUUUGG-CAGUAACUGUUACUGCCG----GCUGCUUUGUUCUGCGGGAUUAGCUGUGAGGCAAUGUCGUGCGAAUUGGCCCGACUUAUGCAGAAUUCUGGGCAAUAACGAUAAUGCAGC- ...(((-(((((.....))))))))----(((((.((((..(((((......)))))..))))((((((....((((.((.((.((......)).)).)).)))).))))))..)))))- ( -38.60, z-score = -1.45, R) >droAna3.scaffold_13417 5902499 102 + 6960332 UUUUGGCCAGAAACUUUGCCUGCCAAACUGUUACUUUGUUCUA-AGGAU--ACUCUGAAG-GAUCUAGGAC-------CUCCACUCUCACGAA-------GCUAUAACUAUAAUGCAACC .((((((.((.........))))))))..((((((((((....-.((((--.((....))-.)))).((..-------..))......)))))-------)...))))............ ( -14.80, z-score = 1.04, R) >droEre2.scaffold_4690 14501993 114 + 18748788 UUUUGG-CAGUAACUGUUACUGCCG----GCUGCUUUGUUCUGCGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGGCUUAUGCAGAAUUCCGGGCAAUAACGAUAAUGCAGC- ...(((-(((((.....))))))))----(((((.(((((.((((.(((..(((....)))...))).)))).....((((((...((.....)).))))))......))))).)))))- ( -43.30, z-score = -1.97, R) >droYak2.chrX 10528046 114 + 21770863 UUUUGG-CAGUAACUGUUACUGCCG----GCUGCUUUGUUCCACGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGACUUAUGCUGAAUUCUGGGCAAUAACGAUAAUGCAGC- ...(((-(((((.....))))))))----(((((.(((((((.((((((((((.(((.(((.(..((....))..).))).)))....)))).))))))))....)))))....)))))- ( -41.90, z-score = -2.59, R) >droSec1.super_15 1583473 114 - 1954846 UUUUGG-CAGUAACUGUUACUGCCG----GCUGCUUUGUUCUACGGGAUUAGCUGUCAGGCGAUAUCGUGCGAAUUGGCUCGACUUAUGCAGAAUUCUGGGCAAUAACGAUAAUGCAGC- ....((-(((((.....)))))))(----((.(((..((((....)))).))).)))..(((.((((((....((((.((.((.((......)).)).)).)))).)))))).)))...- ( -36.70, z-score = -1.74, R) >droSim1.chrX 8434774 114 - 17042790 UUUUGG-CAGUAACUGUUACUGCCG----GCUGCUUUGUUCUACGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGACUUAUGCAGAAUUCUGGGCAAUAACGAUAAUGCAGC- ....((-(((((.....)))))))(----((.(((..((((....)))).))).)))..(((.((((((....((((.((.((.((......)).)).)).)))).)))))).)))...- ( -38.60, z-score = -1.71, R) >consensus UUUUGG_CAGUAACUGUUACUGCCG____GCUGCUUUGUUCUACGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGACUUAUGCAGAAUUCUGGGCAAUAACGAUAAUGCAGC_ ....((.(((((.....))))))).....(((((.(((((..((((......))))..)))))((((((........(((((...............)))))....))))))..))))). (-25.90 = -26.21 + 0.31)

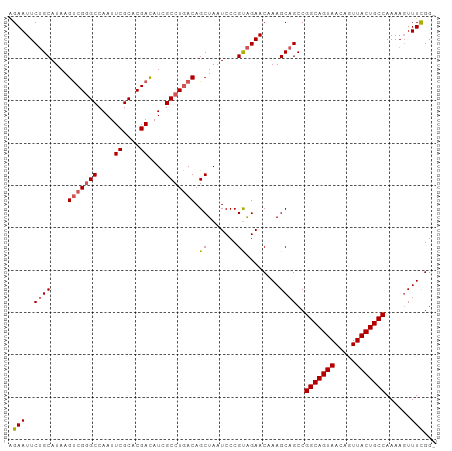
| Location | 10,881,496 – 10,881,596 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.02 |
| Shannon entropy | 0.08824 |
| G+C content | 0.50299 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10881496 100 + 22422827 AGAAUUCUGCAUAAGUCGGGCCAAUUCGCACGACAUUGCCUCACAGCUAAUCCCGCAGAACAAAGCAGCCGGCAGUAACAGUUACUGCCAAAAGUUUCGG- .(((((((((....((((.((......)).))))...((......)).......))))))..........(((((((.....)))))))......)))..- ( -28.20, z-score = -2.28, R) >droEre2.scaffold_4690 14502014 100 - 18748788 GGAAUUCUGCAUAAGCCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGCAGAACAAAGCAGCCGGCAGUAACAGUUACUGCCAAAAGUUUCGG- ((((((((((((.((((((((....((....))....)))))...))).))...)))))...........(((((((.....)))))))....)))))..- ( -27.70, z-score = -1.20, R) >droYak2.chrX 10528067 101 - 21770863 AGAAUUCAGCAUAAGUCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGUGGAACAAAGCAGCCGGCAGUAACAGUUACUGCCAAAAGUUUUGGG .......(((....(((((((....((....))....))))))).)))..(((...))).((((((....(((((((.....)))))))....)))))).. ( -29.50, z-score = -1.67, R) >droSec1.super_15 1583494 100 + 1954846 AGAAUUCUGCAUAAGUCGAGCCAAUUCGCACGAUAUCGCCUGACAGCUAAUCCCGUAGAACAAAGCAGCCGGCAGUAACAGUUACUGCCAAAACUUUCGG- .(((((((((....((((.((......)).))))...((......)).......))))))..........(((((((.....)))))))......)))..- ( -24.30, z-score = -1.61, R) >droSim1.chrX 8434795 100 + 17042790 AGAAUUCUGCAUAAGUCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGUAGAACAAAGCAGCCGGCAGUAACAGUUACUGCCAAAAGUUUCGG- .(((..((((....(((((((....((....))....)))))))..(((......)))......))))..(((((((.....)))))))......)))..- ( -29.50, z-score = -2.53, R) >consensus AGAAUUCUGCAUAAGUCGGGCCAAUUCGCACGACAUCGCCUGACAGCUAAUCCCGUAGAACAAAGCAGCCGGCAGUAACAGUUACUGCCAAAAGUUUCGG_ .(((..((((....(((((((....((....))....))))))).((.......))........))))..(((((((.....)))))))......)))... (-24.92 = -25.32 + 0.40)

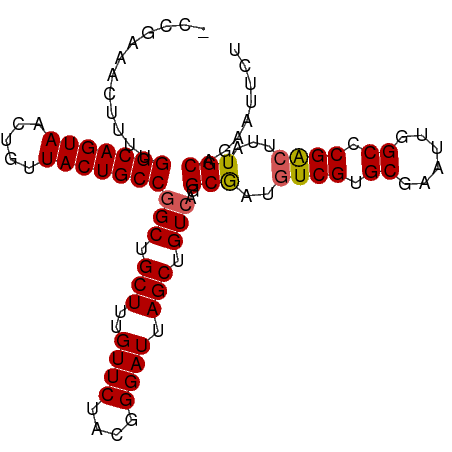
| Location | 10,881,496 – 10,881,596 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Shannon entropy | 0.08824 |
| G+C content | 0.50299 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10881496 100 - 22422827 -CCGAAACUUUUGGCAGUAACUGUUACUGCCGGCUGCUUUGUUCUGCGGGAUUAGCUGUGAGGCAAUGUCGUGCGAAUUGGCCCGACUUAUGCAGAAUUCU -..(((....(((((((((.....)))))))))((((.((((..(((((......)))))..)))).((((.((......)).))))....))))..))). ( -32.50, z-score = -1.48, R) >droEre2.scaffold_4690 14502014 100 + 18748788 -CCGAAACUUUUGGCAGUAACUGUUACUGCCGGCUGCUUUGUUCUGCGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGGCUUAUGCAGAAUUCC -.((((.(..(((((((((.....)))))))))..).))))(((((((.....(((((...(((.(..((....))..).))))))))..))))))).... ( -32.90, z-score = -0.83, R) >droYak2.chrX 10528067 101 + 21770863 CCCAAAACUUUUGGCAGUAACUGUUACUGCCGGCUGCUUUGUUCCACGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGACUUAUGCUGAAUUCU ............(((((((.....)))))))(((.(((..((((....)))).))).))).((((..((((.((......)).))))...))))....... ( -31.90, z-score = -1.55, R) >droSec1.super_15 1583494 100 - 1954846 -CCGAAAGUUUUGGCAGUAACUGUUACUGCCGGCUGCUUUGUUCUACGGGAUUAGCUGUCAGGCGAUAUCGUGCGAAUUGGCUCGACUUAUGCAGAAUUCU -.(....)....(((((((.....)))))))(((.(((..((((....)))).))).)))..((((..(((.((......)).)))..).)))........ ( -29.90, z-score = -0.95, R) >droSim1.chrX 8434795 100 - 17042790 -CCGAAACUUUUGGCAGUAACUGUUACUGCCGGCUGCUUUGUUCUACGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGACUUAUGCAGAAUUCU -..(((......(((((((.....)))))))(((.(((..((((....)))).))).)))..(((..((((.((......)).))))...)))....))). ( -32.80, z-score = -1.79, R) >consensus _CCGAAACUUUUGGCAGUAACUGUUACUGCCGGCUGCUUUGUUCUACGGGAUUAGCUGUCAGGCGAUGUCGUGCGAAUUGGCCCGACUUAUGCAGAAUUCU ............(((((((.....)))))))(((.(((..((((....)))).))).)))..(((..((((.((......)).))))...)))........ (-29.70 = -29.78 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:49 2011