
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,862,156 – 10,862,293 |
| Length | 137 |
| Max. P | 0.889976 |

| Location | 10,862,156 – 10,862,259 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.28 |
| Shannon entropy | 0.23916 |
| G+C content | 0.46616 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.35 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10862156 103 + 22422827 CCGAGAUCCGAGCGUCU----------UCACAGACCCCCAGCAUU------UAUAUUGAGUUGUUGAUUGGGUCCUGAGCCCAAAUGCUUACACGCGAGUGUUUCCUUAACUAAUUAGA ..((((..(..((((..----------.....(((((.(((((..------..........)))))...))))).(((((......))))).))))..)..)))).............. ( -24.40, z-score = -1.58, R) >droSim1.chrX 8415283 103 + 17042790 CCGAGAUCCGAGCGUCU----------UCACAGACCCCCAGCAUU------UAUAUUGAGUUGUUGGUUGGGUCCUGAGCCCAAAUGCUUACACGCGAGGAUUUCCUUAACUAAUUAGA ..(((((((..((((..----------.....(((((((((((..------..........))))))..))))).(((((......))))).))))..))))))).............. ( -33.90, z-score = -3.85, R) >droSec1.super_15 1564498 103 + 1954846 CCGAGAUGCGAGCGUCU----------UCACAGACCCCGAGCAUU------UAUAUUGAGUUGUUGGUUGGGUCCUGAGGCCAAAUGCUUACACGCGAGGAUUUCCUUAACUAAUUAGA ..(((((.(..((((..----------...(((..(((((.((..------.((.....))...)).)))))..)))((((.....))))..))))..).))))).............. ( -25.90, z-score = -0.42, R) >droYak2.chrX 10508509 109 - 21770863 CCGAGAUCCGAGCGUCU----------UAGCAGAUUCCCAGCAUACAUAUGUAUAUUAAGUUGUUGUUUGGGUCCUAAGCCCAAAUGCUUACGCGCGAGUGUUUCCUAAACUAAUUAGA ..((((..(..((((..----------.((((......(((((.((.((.......)).)))))))(((((((.....)))))))))))...))))..)..))))((((.....)))). ( -27.00, z-score = -1.81, R) >droEre2.scaffold_4690 14479760 112 - 18748788 CCGAGAUUCGAGCGAUUCUGCGAGGACUCGCAGAUCCCCAGCACA------UAUAUUGAGUUGUUGUUUGGGUCCUAAGCCCAAAUGCUUACACGCGAGGGUUUCCUAAACUAAUUAG- ..(((((((..(((..(((((((....)))))))....((((.((------.....)).))))..((((((((.....)))))))).......)))..))))))).............- ( -35.30, z-score = -2.71, R) >consensus CCGAGAUCCGAGCGUCU__________UCACAGACCCCCAGCAUU______UAUAUUGAGUUGUUGGUUGGGUCCUGAGCCCAAAUGCUUACACGCGAGGGUUUCCUUAACUAAUUAGA ..(((((.(..((((.................(((((.(((((..................)))))...))))).(((((......))))).))))..).))))).............. (-20.75 = -20.35 + -0.40)

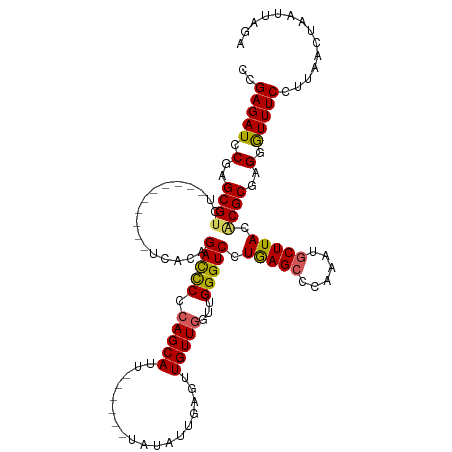

| Location | 10,862,186 – 10,862,293 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Shannon entropy | 0.50876 |
| G+C content | 0.47229 |
| Mean single sequence MFE | -30.01 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.99 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10862186 107 + 22422827 ------GCAUUUAUAUUGAGUUGUUGAUUGGGUC-CUGAGCCCAAAUGCUUACACGCGAGUGUUUCCUUAACUAAUUAGAGAAAUGCUAACCGAAGGAAGUCCACAGCAGCCGU ------...........(.(((((((.((((((.-....))))))......((((....))))((((((......(((((....).))))...)))))).....)))))))).. ( -27.20, z-score = -1.45, R) >droSim1.chrX 8415313 107 + 17042790 ------GCAUUUAUAUUGAGUUGUUGGUUGGGUC-CUGAGCCCAAAUGCUUACACGCGAGGAUUUCCUUAACUAAUUAGAGAAAUGCUCCCCGAAGGAAGUCCACAGCAGCCGG ------..........((.(((((((.((((((.-....)))))).(((......))).((((((((((.........(((.....)))....)))))))))).))))))))). ( -31.72, z-score = -1.82, R) >droSec1.super_15 1564528 107 + 1954846 ------GCAUUUAUAUUGAGUUGUUGGUUGGGUC-CUGAGGCCAAAUGCUUACACGCGAGGAUUUCCUUAACUAAUUAGAGAAAUGCUCCCCGAAGGAAGUCCACAGCAGCCGU ------...........(.(((((((....((((-....))))...(((......))).((((((((((.........(((.....)))....)))))))))).)))))))).. ( -28.72, z-score = -0.75, R) >droYak2.chrX 10508539 113 - 21770863 GCAUACAUAUGUAUAUUAAGUUGUUGUUUGGGUC-CUAAGCCCAAAUGCUUACGCGCGAGUGUUUCCUAAACUAAUUAGAGAAAUGCUCUCCGAAGGAAGUCCACAGCAGCCGU ..((((....)))).....((((((((((((((.-....)))))).(.(((.((.(.(((((((((((((.....)))).))))))))).)))))).).....))))))))... ( -33.30, z-score = -2.91, R) >droEre2.scaffold_4690 14479800 92 - 18748788 ------GCACAUAUAUUGAGUUGUUGUUUGGGUC-CUAAGCCCAAAUGCUUACACGCGAGGGUUUCCUAAACUAAUUAGAG---------------GAAGUCCACAGCAGCCGU ------...........(.((((((((((((((.-....)))))).(((......))).((..(((((...........))---------------)))..))))))))))).. ( -27.30, z-score = -2.01, R) >droAna3.scaffold_13417 5871301 91 - 6960332 -------------UCUUUA--UGUUGGAGGGGUCACGGGGGUCAGGCCCUUA-------AGGCGCCCUAAACUACUAAGAACACGGCUACCGAGGACUCGUCC-CAACAGCCAU -------------......--(((((((.(((((.(((..(((.(((((...-------.)).))).....((....)).....)))..)))..))))).).)-)))))..... ( -31.80, z-score = -1.11, R) >consensus ______GCAUUUAUAUUGAGUUGUUGGUUGGGUC_CUGAGCCCAAAUGCUUACACGCGAGGGUUUCCUAAACUAAUUAGAGAAAUGCUCCCCGAAGGAAGUCCACAGCAGCCGU ...................(((((((.((((((......))))))..............(((((((((..........................))))))))).)))))))... (-15.68 = -16.99 + 1.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:45 2011