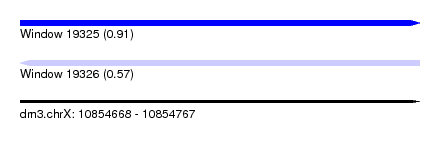
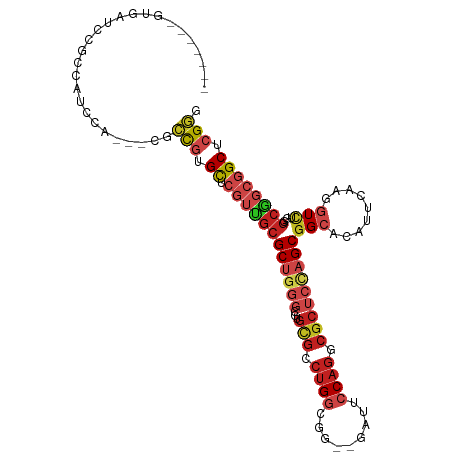
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,854,668 – 10,854,767 |
| Length | 99 |
| Max. P | 0.912488 |

| Location | 10,854,668 – 10,854,767 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.59421 |
| G+C content | 0.69081 |
| Mean single sequence MFE | -44.78 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
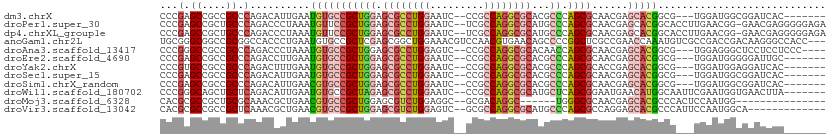
>dm3.chrX 10854668 99 + 22422827 CCCGAGCCGCCGCCCAGACAUUGAAUGUGCCGCUGGAGCGCCUGGAAUC--CCGCCAGGCGCACGCCCAGCGCAACGAGCACGGCG---UGGAUGGCGGAUCAC------- ...((.((((((.(((.((((...))))((((((((.((((((((....--...))))))))....)))))((.....))..))).---))).)))))).))..------- ( -49.20, z-score = -3.25, R) >droPer1.super_30 422937 108 - 958394 CCCGAGCCGCUGCCCAGACCCUAAAUGUUCCGCUGGAGCGCCUGGAAUC--UCGCCAGGCGCAUGCCCAGCGCAACGAGCACGGCACCUUGAACGG-GAACGAGGGGGAGA (((..((((.(((.(((...))....(((.((((((.((((((((....--...))))))))....)))))).)))).))))))).(((((.....-...))))))))... ( -48.20, z-score = -2.09, R) >dp4.chrXL_group1e 680432 108 + 12523060 CCCGAGCCGCUGCCCAGACCCUAAAUGUUCCGCUGGAGCGCCUGGAAUC--UCGCCAGGCGCAUGCCCAGCGCAACGAGCACGGCACCUUGAACGG-GAACGAGGGGGAGA (((..((((.(((.(((...))....(((.((((((.((((((((....--...))))))))....)))))).)))).))))))).(((((.....-...))))))))... ( -48.20, z-score = -2.09, R) >anoGam1.chr2L 36995060 108 - 48795086 UGCGGGCGGCCGCGGCCACCCUGAAUGUGCCGCUCGAGCGGCUGGAAACGUCCAACGUGAACAGCCCCGGCUCGCCGAACCAAAUGUCGCCGACCGACAAGGGCCACC--- ....((((((.(((((((.......)).)))))..(((((((((...(((.....)))...)))))...)))))))...((...(((((.....))))).)))))...--- ( -44.40, z-score = -0.58, R) >droAna3.scaffold_13417 5864165 102 - 6960332 CCCGGGCCGCCGCCCAGACCCUAAAUGUGCCGCUGGAGCGCCUGGAGUC--CCGCCAGGCGCACAACCAGCGCAACGAGCACGGCG---UGGAGGGCUCCUCCUCCC---- ...((((....)))).............((((((((.((((((((....--...))))))))....)))))((.....))..))).---.((((((....)))))).---- ( -47.40, z-score = -1.50, R) >droEre2.scaffold_4690 14472758 99 - 18748788 CCCGAGCCGCCGCCCAGACCUUGAAUGUGCCGCUGGAGCGCCUGGAAUC--CCGCCAGGCGCACGCCCAGCGCAACGAGCACGGCG---UGGAUGGGGGAUUGC------- (((.(.((((.(((.....((((....(((.(((((.((((((((....--...))))))))....)))))))).))))...))))---))).).)))......------- ( -46.00, z-score = -1.47, R) >droYak2.chrX 10501245 99 - 21770863 CCCGUGCCGCCGCCCAGACUUUGAAUGUGCCGCUGGAGCGCCUGGAAUC--CCGCCAGGCGCACGCCCAGCGCACCGAGCACGGCG---UGGAUGGAGGAUCAC------- .((((.((((.(((..(.(((.....((((.(((((.((((((((....--...))))))))....))))))))).))))..))))---)))))))........------- ( -44.10, z-score = -1.64, R) >droSec1.super_15 1556976 99 + 1954846 CCCGAGCCGCCGCCCAGACAUUGAAUGUGCCGCUGGAGCGCCUGGAAUC--CCGCCAGGCGCACGCCCAGCGCAACGAGCACGGCG---UGGAUGGCGGAUCAC------- ...((.((((((.(((.((((...))))((((((((.((((((((....--...))))))))....)))))((.....))..))).---))).)))))).))..------- ( -49.20, z-score = -3.25, R) >droSim1.chrX_random 3059087 99 + 5698898 CCCGAGCCGCCGCCCAGACAUUGAACGUGCCGCUGGAGCGCCUGGAAUC--CCGCCAGGCGCACGCCCAGCGCAACGAGCACGGCG---UGGAUGGCGGAUCAC------- ...((.((((((.(((..(..((..(((..((((((.((((((((....--...))))))))....))))))..)))..))..)..---))).)))))).))..------- ( -48.70, z-score = -3.02, R) >droWil1.scaffold_180702 904392 102 + 4511350 CCCGGGCAGCUGCUCAGACAUUGAAUGUGCCGCUAGAGCGCCUGGAAUC--CCGCCAGGCGCAUGCUCAGCGGAAUGAACAUGGCAAUUCGAAUGGUGAACUUA------- .((((((....)))).((.((((.((((.(((((((.((((((((....--...))))))))...)).))))).....))))..))))))....))........------- ( -38.30, z-score = -1.41, R) >droMoj3.scaffold_6328 695767 88 - 4453435 CACGCGCCGCUGCGCAAACGCUGAACGUGCCGCUGGAGCGUCUGGAGGC--GCGACAGGC------UGGGCGCAACGAGCACGCCCACUCCAAUGG--------------- ..((.((((.(((((....(((...((((((.(..(.....)..).)))--)))...)))------...))))).)).)).)).(((......)))--------------- ( -35.70, z-score = 0.52, R) >droVir3.scaffold_13042 1895729 96 - 5191987 CACGCGCCGCCGCUCAAACGCUGAACGUGCCGCUGGAGCGUCUGGAGUC--GCGCCAGGCGCAUGCCCAGCGCCAGGAGCACGCCCAUUCCAAUGGCA------------- ...........((((...(((((..((((((.((((.(((.(....).)--)).))))).)))))..)))))....))))..(((.........))).------------- ( -38.00, z-score = 0.68, R) >consensus CCCGAGCCGCCGCCCAGACCCUGAAUGUGCCGCUGGAGCGCCUGGAAUC__CCGCCAGGCGCACGCCCAGCGCAACGAGCACGGCG___UGGAUGGCGGAUCAC_______ ...(.((....)).)..........(((((((((((.((((((((.........))))))))...)).))))......)))))............................ (-20.70 = -21.42 + 0.72)

| Location | 10,854,668 – 10,854,767 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.59421 |
| G+C content | 0.69081 |
| Mean single sequence MFE | -47.35 |
| Consensus MFE | -25.16 |
| Energy contribution | -26.48 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
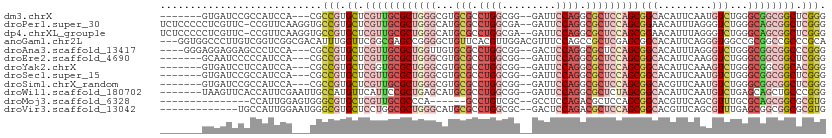
>dm3.chrX 10854668 99 - 22422827 -------GUGAUCCGCCAUCCA---CGCCGUGCUCGUUGCGCUGGGCGUGCGCCUGGCGG--GAUUCCAGGCGCUCCAGCGGCACAUUCAAUGUCUGGGCGGCGGCUCGGG -------.(((.(((((..(((---.(((..((.....))((((((...((((((((...--....)))))))))))))))))((((...)))).)))..))))).))).. ( -49.10, z-score = -1.01, R) >droPer1.super_30 422937 108 + 958394 UCUCCCCCUCGUUC-CCGUUCAAGGUGCCGUGCUCGUUGCGCUGGGCAUGCGCCUGGCGA--GAUUCCAGGCGCUCCAGCGGAACAUUUAGGGUCUGGGCAGCGGCUCGGG .....(((......-((......)).((((((((((((.(((((((...((((((((...--....))))))))))))))).)))....((...))))))).))))..))) ( -48.30, z-score = -0.77, R) >dp4.chrXL_group1e 680432 108 - 12523060 UCUCCCCCUCGUUC-CCGUUCAAGGUGCCGUGCUCGUUGCGCUGGGCAUGCGCCUGGCGA--GAUUCCAGGCGCUCCAGCGGAACAUUUAGGGUCUGGGCAGCGGCUCGGG .....(((......-((......)).((((((((((((.(((((((...((((((((...--....))))))))))))))).)))....((...))))))).))))..))) ( -48.30, z-score = -0.77, R) >anoGam1.chr2L 36995060 108 + 48795086 ---GGUGGCCCUUGUCGGUCGGCGACAUUUGGUUCGGCGAGCCGGGGCUGUUCACGUUGGACGUUUCCAGCCGCUCGAGCGGCACAUUCAGGGUGGCCGCGGCCGCCCGCA ---.((((((..(((((.....)))))...)))..((((.((((.(((..(((....((((....))))(((((....))))).......)))..))).))))))))))). ( -54.00, z-score = -1.30, R) >droAna3.scaffold_13417 5864165 102 + 6960332 ----GGGAGGAGGAGCCCUCCA---CGCCGUGCUCGUUGCGCUGGUUGUGCGCCUGGCGG--GACUCCAGGCGCUCCAGCGGCACAUUUAGGGUCUGGGCGGCGGCCCGGG ----(((.(((((...))))).---((((((((((..((((((((....((((((((...--....)))))))).))))).)))......))))....)))))).)))... ( -53.40, z-score = -0.60, R) >droEre2.scaffold_4690 14472758 99 + 18748788 -------GCAAUCCCCCAUCCA---CGCCGUGCUCGUUGCGCUGGGCGUGCGCCUGGCGG--GAUUCCAGGCGCUCCAGCGGCACAUUCAAGGUCUGGGCGGCGGCUCGGG -------...............---..(((.((.((((((((((((...((((((((...--....))))))))))))))(((.(......))))...)))))))).))). ( -44.10, z-score = 0.42, R) >droYak2.chrX 10501245 99 + 21770863 -------GUGAUCCUCCAUCCA---CGCCGUGCUCGGUGCGCUGGGCGUGCGCCUGGCGG--GAUUCCAGGCGCUCCAGCGGCACAUUCAAAGUCUGGGCGGCGGCACGGG -------(((..........))---).((((((.((.(((((((((...((((((((...--....))))))))))))))(((.........)))...))).)))))))). ( -46.60, z-score = -0.54, R) >droSec1.super_15 1556976 99 - 1954846 -------GUGAUCCGCCAUCCA---CGCCGUGCUCGUUGCGCUGGGCGUGCGCCUGGCGG--GAUUCCAGGCGCUCCAGCGGCACAUUCAAUGUCUGGGCGGCGGCUCGGG -------.(((.(((((..(((---.(((..((.....))((((((...((((((((...--....)))))))))))))))))((((...)))).)))..))))).))).. ( -49.10, z-score = -1.01, R) >droSim1.chrX_random 3059087 99 - 5698898 -------GUGAUCCGCCAUCCA---CGCCGUGCUCGUUGCGCUGGGCGUGCGCCUGGCGG--GAUUCCAGGCGCUCCAGCGGCACGUUCAAUGUCUGGGCGGCGGCUCGGG -------.(((.(((((..(((---.((..((..(((.((((((((...((((((((...--....)))))))))))))).)))))..))..)).)))..))))).))).. ( -50.60, z-score = -1.13, R) >droWil1.scaffold_180702 904392 102 - 4511350 -------UAAGUUCACCAUUCGAAUUGCCAUGUUCAUUCCGCUGAGCAUGCGCCUGGCGG--GAUUCCAGGCGCUCUAGCGGCACAUUCAAUGUCUGAGCAGCUGCCCGGG -------...(((((.((((.(((((((((((((((......)))))))((((((((...--....))))))))......)))).))))))))..)))))........... ( -38.90, z-score = -1.70, R) >droMoj3.scaffold_6328 695767 88 + 4453435 ---------------CCAUUGGAGUGGGCGUGCUCGUUGCGCCCA------GCCUGUCGC--GCCUCCAGACGCUCCAGCGGCACGUUCAGCGUUUGCGCAGCGGCGCGUG ---------------((((....))))((((((.((((((((..(------(((((.((.--(((.(..(.....)..).))).))..))).))).)))))))))))))). ( -39.70, z-score = -0.26, R) >droVir3.scaffold_13042 1895729 96 + 5191987 -------------UGCCAUUGGAAUGGGCGUGCUCCUGGCGCUGGGCAUGCGCCUGGCGC--GACUCCAGACGCUCCAGCGGCACGUUCAGCGUUUGAGCGGCGGCGCGUG -------------..((((....))))(((((((.(((((((..(((....)))..))))--..(((.(((((((..((((...)))).))))))))))))).))))))). ( -46.10, z-score = -0.22, R) >consensus _______GUGAUCCGCCAUCCA___CGCCGUGCUCGUUGCGCUGGGCGUGCGCCUGGCGG__GAUUCCAGGCGCUCCAGCGGCACAUUCAAGGUCUGGGCGGCGGCUCGGG ...........................(((.((.((((((((((((...(((.((((.(.....).)))).)))))))))(((.........)))...)))))))).))). (-25.16 = -26.48 + 1.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:43 2011