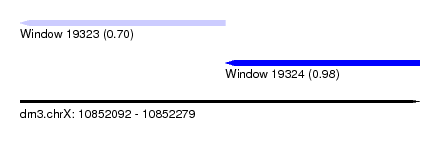
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,852,092 – 10,852,279 |
| Length | 187 |
| Max. P | 0.981055 |

| Location | 10,852,092 – 10,852,188 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.53 |
| Shannon entropy | 0.07881 |
| G+C content | 0.41634 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -21.24 |
| Energy contribution | -20.92 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
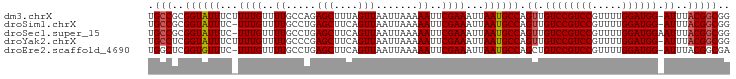
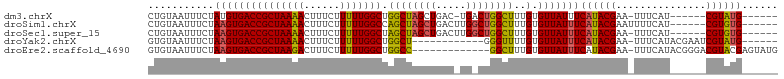
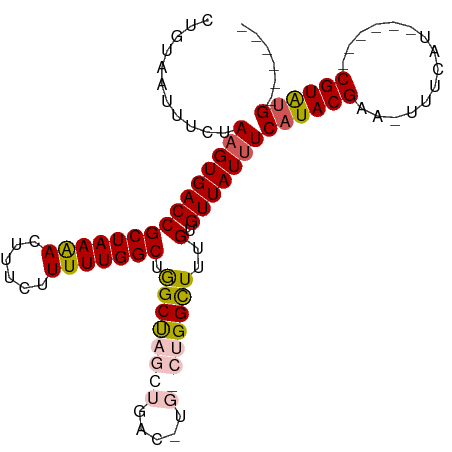
>dm3.chrX 10852092 96 - 22422827 UGCCGCGGUAUUUCUUUUGUUUUGCCAGAGCUUUAGUUAAUUAAAAAUUCGAAAUUAAUGCCAGUUGUCCGUCCGUUUUGGAUGG-AUUUACGGCGG ..((((((((((..(((((..((.....(((....))).......))..)))))..)))))).((.((((((((.....))))))-))..)).)))) ( -29.80, z-score = -3.02, R) >droSim1.chrX 8405444 95 - 17042790 UGCCGCGGUAUUUC-UUUGUUUUGCCUGAGCUUCAGUUAAUUAAAAAUUCGAAAUUAAUGCCAGUUGUCCGUCCGUUUUGGAUGG-AUUUACGGCGG ..((((((((((..-((((..((.....(((....))).......))..))))...)))))).((.((((((((.....))))))-))..)).)))) ( -27.50, z-score = -2.20, R) >droSec1.super_15 1554405 96 - 1954846 UGCCGCGGUAUUUC-UUUGUUUUGCCUGAGCUUCAGUUAAUUAAAAAUUCGAAAUUAAUGCCAGUUGUCCGUCCGUUUUGGAUGGAAUUUACGGCGG ..((((((((((..-((((..((.....(((....))).......))..))))...)))))).((..(((((((.....)))))))....)).)))) ( -26.20, z-score = -1.74, R) >droYak2.chrX 10498673 96 + 21770863 UGCCUCGGUAUUUCUUUUGUUUUGCCCGAGCUUCAGUUAAUUAAAAAUUCGAAAUUAAUGCCAGUUGUCCGUCCGUUUUGGAUGG-AUUUACGGCGG .(((..((((((..(((((..((.....(((....))).......))..)))))..)))))).((.((((((((.....))))))-))..))))).. ( -28.70, z-score = -2.88, R) >droEre2.scaffold_4690 14470288 95 + 18748788 UGGCUCGGUGUUUC-UUUGUUUUGCCUGAGCUUCAGUUAAUUAAAAAUUCGAAAUUAAUGCCAGCUGUCCGUCCGUUUUGGAUGG-AUUUACGGCGA .(((((((.((...-........)))))))))...(((((((..........)))))))(((....((((((((.....))))))-))....))).. ( -27.70, z-score = -2.19, R) >consensus UGCCGCGGUAUUUC_UUUGUUUUGCCUGAGCUUCAGUUAAUUAAAAAUUCGAAAUUAAUGCCAGUUGUCCGUCCGUUUUGGAUGG_AUUUACGGCGG ......((((((...((((..((.....(((....))).......))..))))...)))))).(((((((((((.....)))))).....))))).. (-21.24 = -20.92 + -0.32)
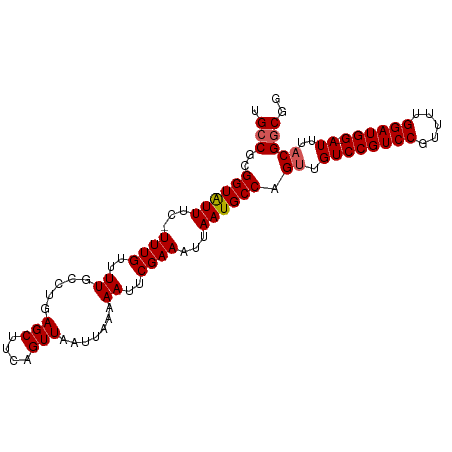
| Location | 10,852,188 – 10,852,279 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Shannon entropy | 0.31330 |
| G+C content | 0.39036 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.981055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10852188 91 - 22422827 CUGUAAUUUCUAUGUGACCGCUAAAACUUUCUUUUUGGCUGGCUAGCUGAC-UGACUGGCUUUGUGUUAUUUCAUACGAA-UUUCAU------CGUAUG------ .............(((((((((((((......))))))).(((((((....-.).))))))..).)))))..(((((((.-.....)------))))))------ ( -20.80, z-score = -2.90, R) >droSim1.chrX 8405539 93 - 17042790 CUGUAAUUUCUAAGUGACCGCUAAAACUUUCUUUUUGGCCAGCUAGCUGACUUGGCUGGCUUUGUGUUAUUUCAUACGAAUUUUCAU------CGUGUG------ ...........(((((((((((((((......))))))).((((((((.....))))))))..).)))))))(((((((.......)------))))))------ ( -26.10, z-score = -3.44, R) >droSec1.super_15 1554501 92 - 1954846 CUGUAAUUUCUAAGUGACCGCUAAAACUUUCUUUUUGGCUAGCUAGCUGACUUGGCUGGCUUUGUGUUAUUUCAUACGAA-UUUCAU------CGUGUG------ ...........(((((((((((((((......))))))).((((((((.....))))))))..).)))))))(((((((.-.....)------))))))------ ( -26.90, z-score = -4.07, R) >droYak2.chrX 10498769 86 + 21770863 GUGUAAUUUCUAAGUGACCGCUAAAACUUUCUUUUUGGCUGGCU------------GGGUUUUGUGUUAUUUCAUACGAA-UUUCAUACGAAUCGUAUG------ (..(((..((((.((.(..(((((((......)))))))).)))------------)))..)))..).....(((((((.-.(((....))))))))))------ ( -16.50, z-score = -1.11, R) >droEre2.scaffold_4690 14470383 91 + 18748788 GUGUAAUUUCUAAGUGACCGCUAAGACUUUCUUUUUGGCUGGCC-------------GGCUUUGUGUUAUUUCAUACGAA-UUUCAUACGGGACGUACGAGUAUG .((((..((((..(((((((((((((......))))))).))..-------------...(((((((......)))))))-..))))..))))..))))...... ( -16.80, z-score = 0.64, R) >consensus CUGUAAUUUCUAAGUGACCGCUAAAACUUUCUUUUUGGCUGGCUAGCUGAC_UG_CUGGCUUUGUGUUAUUUCAUACGAA_UUUCAU______CGUAUG______ ...........(((((((((((((((......))))))).((((((((.....))))))))..).)))))))((((((...............))))))...... (-17.69 = -17.88 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:41 2011