| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,851,640 – 10,851,733 |
| Length | 93 |
| Max. P | 0.825628 |

| Location | 10,851,640 – 10,851,733 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.39761 |
| G+C content | 0.48555 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -13.01 |
| Energy contribution | -14.49 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
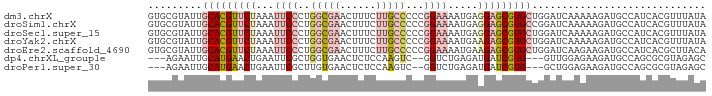
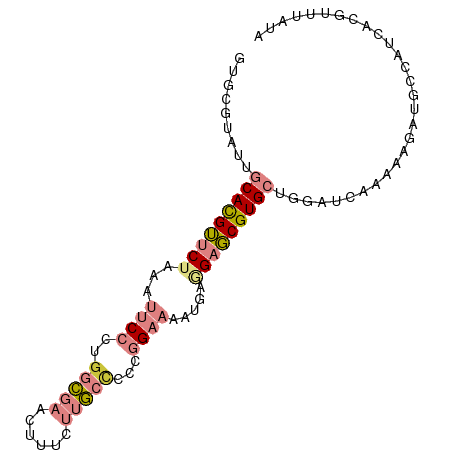
>dm3.chrX 10851640 93 + 22422827 GUGCGUAUUGCACGUUCUAAAUUCCCUGGCGAACUUUCUUGCCCCCGGAAAAUGAGGAGCGUGCUGGAUCAAAAAGAUGCCAUCACGUUUAUA ..((((...(((((((((...((((..(((((......)))))...)))).....)))))))))((((((.....))).)))..))))..... ( -29.40, z-score = -2.77, R) >droSim1.chrX 8405008 93 + 17042790 GUGCGUAUUGCACGUUCUAAAUUCCCUGGCGAACUUUCUUGCCCCCGGAAAAUGAGGAGCGUGCCGGAUCAAAAAGAUGCCAUCACGUUUAUA ..((((...(((((((((...((((..(((((......)))))...)))).....))))))))).(((((.....))).))...))))..... ( -28.70, z-score = -2.41, R) >droSec1.super_15 1553958 93 + 1954846 GUGCGUAUUGCACGUUCUAAAUUCCCUGGCGAACUUUCUUGCCCCCGGAAAAUGAGGAGCGUGCUGGAUCAAAAAGAUGCCAUCACGUUUAUA ..((((...(((((((((...((((..(((((......)))))...)))).....)))))))))((((((.....))).)))..))))..... ( -29.40, z-score = -2.77, R) >droYak2.chrX 10498250 93 - 21770863 GUGCGUAUUGCACGUUCUAAAUUCCCUGGCGAACUUUCUUGCCCCCGGAAAAUGAAGAGCGUGCUGGAUCAAAAAGAUGCCAUCACGUUUAUA ..((((...(((((((((...((((..(((((......)))))...)))).....)))))))))((((((.....))).)))..))))..... ( -29.40, z-score = -3.32, R) >droEre2.scaffold_4690 14469856 93 - 18748788 GUGCGUAUUGCACGUUCUAAAUUCCCUGGCGAACUUUCUUGCCCCCGGAAAAUGAAGAGCGUGCUGGAUCAAGAAGAUGCCAUCACGCUUACA ..((((...(((((((((...((((..(((((......)))))...)))).....)))))))))((((((.....))).)))..))))..... ( -31.70, z-score = -3.40, R) >dp4.chrXL_group1e 677227 85 + 12523060 ---AGAAUUGCAUGAACUGAAUUCGCUGGUGAACUCUCCAAGUC--GGUCUGAGAUGAUCGUG---GUUGGAGAAGAUGCCAGCGCGUAGAGC ---.(((((.((.....)))))))(((((((...(((((((.((--((((......))))).)---.)))))))...)))))))((.....)) ( -30.20, z-score = -2.47, R) >droPer1.super_30 419748 85 - 958394 ---AGAAUUGCAUGAACUGAAUUCGCUUGUGAACUCUCCAAGUC--GGUCUGAGAUGAUCGUG---GCUGGAGAAGAUGCCAGCGCGUAGAGC ---.(((((.((.....)))))))(((.(((...((((((.(((--((((......)))))..---))))))))...))).)))((.....)) ( -22.20, z-score = 0.31, R) >consensus GUGCGUAUUGCACGUUCUAAAUUCCCUGGCGAACUUUCUUGCCCCCGGAAAAUGAGGAGCGUGCUGGAUCAAAAAGAUGCCAUCACGUUUAUA .........(((((((((...((((..(((((......)))))...)))).....)))))))))............................. (-13.01 = -14.49 + 1.47)

| Location | 10,851,640 – 10,851,733 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.39761 |
| G+C content | 0.48555 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -13.57 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
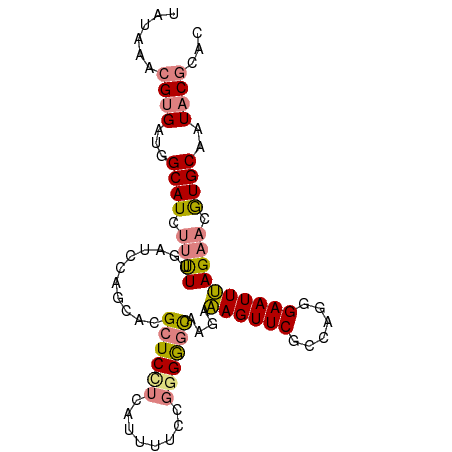
>dm3.chrX 10851640 93 - 22422827 UAUAAACGUGAUGGCAUCUUUUUGAUCCAGCACGCUCCUCAUUUUCCGGGGGCAAGAAAGUUCGCCAGGGAAUUUAGAACGUGCAAUACGCAC .......(((.(((.(((.....))))))(((((.((.....(((((((((((......)))).)).)))))....)).)))))....))).. ( -26.80, z-score = -1.89, R) >droSim1.chrX 8405008 93 - 17042790 UAUAAACGUGAUGGCAUCUUUUUGAUCCGGCACGCUCCUCAUUUUCCGGGGGCAAGAAAGUUCGCCAGGGAAUUUAGAACGUGCAAUACGCAC .......(((.(((.(((.....))))))(((((.((.....(((((((((((......)))).)).)))))....)).)))))....))).. ( -25.90, z-score = -1.18, R) >droSec1.super_15 1553958 93 - 1954846 UAUAAACGUGAUGGCAUCUUUUUGAUCCAGCACGCUCCUCAUUUUCCGGGGGCAAGAAAGUUCGCCAGGGAAUUUAGAACGUGCAAUACGCAC .......(((.(((.(((.....))))))(((((.((.....(((((((((((......)))).)).)))))....)).)))))....))).. ( -26.80, z-score = -1.89, R) >droYak2.chrX 10498250 93 + 21770863 UAUAAACGUGAUGGCAUCUUUUUGAUCCAGCACGCUCUUCAUUUUCCGGGGGCAAGAAAGUUCGCCAGGGAAUUUAGAACGUGCAAUACGCAC .......(((.(((.(((.....))))))(((((.(((..(((..(.((((((......)))).)).)..)))..))).)))))....))).. ( -29.40, z-score = -3.00, R) >droEre2.scaffold_4690 14469856 93 + 18748788 UGUAAGCGUGAUGGCAUCUUCUUGAUCCAGCACGCUCUUCAUUUUCCGGGGGCAAGAAAGUUCGCCAGGGAAUUUAGAACGUGCAAUACGCAC .....(((((.(((.(((.....))))))(((((.(((..(((..(.((((((......)))).)).)..)))..))).)))))..))))).. ( -33.40, z-score = -3.21, R) >dp4.chrXL_group1e 677227 85 - 12523060 GCUCUACGCGCUGGCAUCUUCUCCAAC---CACGAUCAUCUCAGACC--GACUUGGAGAGUUCACCAGCGAAUUCAGUUCAUGCAAUUCU--- ((.....))(((((....((((((((.---..((.((......)).)--)..))))))))....)))))(((((((.....)).))))).--- ( -24.40, z-score = -2.72, R) >droPer1.super_30 419748 85 + 958394 GCUCUACGCGCUGGCAUCUUCUCCAGC---CACGAUCAUCUCAGACC--GACUUGGAGAGUUCACAAGCGAAUUCAGUUCAUGCAAUUCU--- ((.....))(((......((((((((.---..((.((......)).)--)..))))))))......)))(((((((.....)).))))).--- ( -18.70, z-score = -0.12, R) >consensus UAUAAACGUGAUGGCAUCUUUUUGAUCCAGCACGCUCCUCAUUUUCCGGGGGCAAGAAAGUUCGCCAGGGAAUUUAGAACGUGCAAUACGCAC ......((((...((((.((((...........((((((........))))))....((((((......)))))))))).))))..))))... (-13.57 = -14.19 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:40 2011