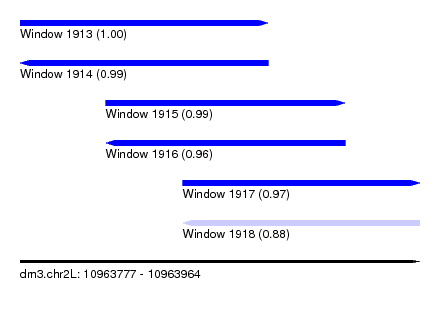
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,963,777 – 10,963,964 |
| Length | 187 |
| Max. P | 0.998297 |

| Location | 10,963,777 – 10,963,893 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 63.50 |
| Shannon entropy | 0.67004 |
| G+C content | 0.39636 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
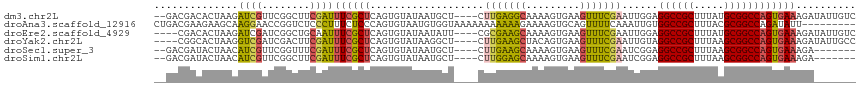
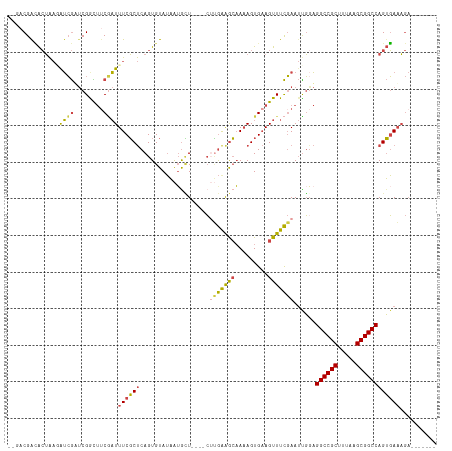
>dm3.chr2L 10963777 116 + 23011544 AGCAGGGCGAAUAUUUCCCAUGAAUUUAACCAUUUAAACAUCCAAUGUUAGUUUUUAACAGACAAUUAAUUCA-GGGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUU ....(((.(((((((((((.((((((..........(((((...))))).((((.....))))....))))))-.)))).)))))...))...((((((.....))))))))).... ( -28.40, z-score = -2.79, R) >droAna3.scaffold_12916 14825715 93 + 16180835 --------GAAUGCAUCC-AUUAAUUUAAU--UUUAA------GACUUCCCACUGCAAUAAGUAUUUCUGUC-------UAGCGACAAUAUCUGGCCGCGUAAAGCGGCCACAAUUU --------...(((....-..(((......--.)))(------(((.....(((......)))......)))-------).)))........(((((((.....)))))))...... ( -19.60, z-score = -1.85, R) >droEre2.scaffold_4929 12156018 100 - 26641161 ---ACGACGAAUAUCUUC-AUAAACUGAAG-------------AAUGUCAGUAUUUUGUUGGUGGUUGAUGCAAGGGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUU ---..((((....(((((-(.....)))))-------------).))))...........(((((..((((...(....)..))))..)))))((((((.....))))))....... ( -30.30, z-score = -2.58, R) >droYak2.chr2L 7355566 113 + 22324452 ---ACGACGAAUAUCUCC-AUAAACGGAAGUAUUCAAGCAUCCAAUGUUAGUAUUUGGUUGGUUGUUGAUACAGGGGGGCAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUACAAUU ---.(((((((((..(((-......)))..)))))((.((.((((.........)))).)).)))))).....(..(((.....)))..)...((((((.....))))))....... ( -34.00, z-score = -2.54, R) >droSec1.super_3 6364586 103 + 7220098 AGCUGGGCGAUUAUCUCCCAUAAAUUAAACCAUUUAAACAUCCAAAGUUAGUUUUUAACAGACAAUUGAUUC--------------UUUCACUGGCCGCUUAAAGCGGCCUCCGAUU ...(((((((((.(((.....(((((((....(((........))).))))))).....))).)))))....--------------.......((((((.....))))))))))... ( -20.80, z-score = -1.41, R) >droSim1.chr2L 10760562 103 + 22036055 AGCUGUGCGAUUAUCUCCCAUAAAUUAAACCAUUUAAACAUCCAAUGUUAGUUUUUAACAGACAAUUGAUUC--------------UUUCACUGGCCGCUUAAAGCGGCCUCCGAUU .......(((((.(((.....(((((((..((((.........))))))))))).....))).)))))....--------------..((...((((((.....))))))...)).. ( -19.40, z-score = -1.69, R) >consensus ___AGGGCGAAUAUCUCC_AUAAAUUAAACCAUUUAAACAUCCAAUGUUAGUUUUUAACAGACAAUUGAUUC_______CAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUCCAAUU .............................................................................................((((((.....))))))....... (-13.20 = -13.20 + 0.00)
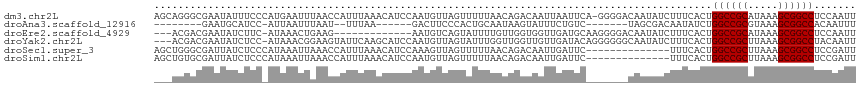
| Location | 10,963,777 – 10,963,893 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.50 |
| Shannon entropy | 0.67004 |
| G+C content | 0.39636 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10963777 116 - 23011544 AAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCCC-UGAAUUAAUUGUCUGUUAAAAACUAACAUUGGAUGUUUAAAUGGUUAAAUUCAUGGGAAAUAUUCGCCCUGCU ....((.((((((.....)))))).((((...(((((.((((.-(((((((((..(...(((((..(((....)))...))))))..))).)))))).))))))))))))))).... ( -33.30, z-score = -2.79, R) >droAna3.scaffold_12916 14825715 93 - 16180835 AAAUUGUGGCCGCUUUACGCGGCCAGAUAUUGUCGCUA-------GACAGAAAUACUUAUUGCAGUGGGAAGUC------UUAAA--AUUAAAUUAAU-GGAUGCAUUC-------- ....(((((((((.....))))))((((.(((((....-------))))).....(((((....)))))..)))------)....--...........-....)))...-------- ( -21.80, z-score = -1.03, R) >droEre2.scaffold_4929 12156018 100 + 26641161 AAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCCCUUGCAUCAACCACCAACAAAAUACUGACAUU-------------CUUCAGUUUAU-GAAGAUAUUCGUCGU--- ..((((.((((((.....)))))).(..(..(((...)))...)..)........)))).........(((..(-------------(((((.....)-))))).....)))..--- ( -26.10, z-score = -2.38, R) >droYak2.chr2L 7355566 113 - 22324452 AAUUGUAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGCCCCCCUGUAUCAACAACCAACCAAAUACUAACAUUGGAUGCUUGAAUACUUCCGUUUAU-GGAGAUAUUCGUCGU--- ..((((.((((((.....)))))).......(((((.........))))).))))....((((.........))))(((..((((((.(((((....)-)))).)))))).)))--- ( -27.90, z-score = -1.80, R) >droSec1.super_3 6364586 103 - 7220098 AAUCGGAGGCCGCUUUAAGCGGCCAGUGAAA--------------GAAUCAAUUGUCUGUUAAAAACUAACUUUGGAUGUUUAAAUGGUUUAAUUUAUGGGAGAUAAUCGCCCAGCU ..(((..((((((.....))))))..)))..--------------((((((.(((..((((.(((......))).))))..))).))))))......((((.(.....).))))... ( -25.00, z-score = -0.90, R) >droSim1.chr2L 10760562 103 - 22036055 AAUCGGAGGCCGCUUUAAGCGGCCAGUGAAA--------------GAAUCAAUUGUCUGUUAAAAACUAACAUUGGAUGUUUAAAUGGUUUAAUUUAUGGGAGAUAAUCGCACAGCU .......((((((.....)))))).(((..(--------------..(((..(..(..((((((....(((((...))))).......))))))..)..)..)))..)..))).... ( -23.60, z-score = -0.94, R) >consensus AAUUGGAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUG_______GAAUCAAUUACCUAUUAAAAACUAACAUUGGAUGUUUAAAUGGUUAAAUUUAU_GGAGAUAUUCGCCCU___ .......((((((.....))))))............................................................................................. (-13.82 = -13.82 + 0.00)
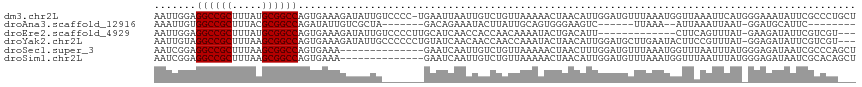
| Location | 10,963,817 – 10,963,929 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.91 |
| Shannon entropy | 0.53251 |
| G+C content | 0.40580 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10963817 112 + 23011544 ---UCCAAUGUUAGUUUUUAACAGACAAUUAAUUCA-GGGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCCUCAAG----AGCAUUAUACACU ---...((((((.((((.....))))..........-((((.(((...........((((((.....))))))........((....))....)))))))...----))))))....... ( -23.30, z-score = -2.15, R) >droAna3.scaffold_12916 14825746 102 + 16180835 CCCACUGCAAUAAGUAUUU--CUGUCUAGCGACAA----------------UAUCUGGCCGCGUAAAGCGGCCACAAUUUGAAAACUGCACUUUUCUUUUUUUUUUUACCACAUUACACU .....((((....((.(((--(((((....)))).----------------....(((((((.....)))))))......)))))))))).............................. ( -21.20, z-score = -2.61, R) >droEre2.scaffold_4929 12156044 110 - 26641161 ------AAUGUCAGUAUUUUGUUGGUGGUUGAUGCAAGGGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUCGCG----AAUAUUAUACACU ------.......((((..((((.((((.....(((((((((..............((((((.....))))))..............)).))))))).)))).----))))..))))... ( -31.69, z-score = -2.57, R) >droYak2.chr2L 7355602 113 + 22324452 ---UCCAAUGUUAGUAUUUGGUUGGUUGUUGAUACAGGGGGGCAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUACAAUUCGAAACUUCACUGUAGCUUCAAG----AGCCUUAUACACU ---.((((.........))))..((((.((((((((((..((......))..)...((((((.....)))))).................)))))...)))).----))))......... ( -31.30, z-score = -1.36, R) >droSec1.super_3 6364626 99 + 7220098 ---UCCAAAGUUAGUUUUUAACAGACAAUUGAUUC--------------UUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUUCAAG----AGCAUUAUACACU ---...(((((.((((((...........(((...--------------..)))..((((((.....))))))........))))))..)))))(((((...)----))))......... ( -21.80, z-score = -2.49, R) >droSim1.chr2L 10760602 99 + 22036055 ---UCCAAUGUUAGUUUUUAACAGACAAUUGAUUC--------------UUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUCCAAG----AGCAUUAUACACU ---..(((((((.((.....)).))).)))).(((--------------..((...((((((.....))))))...))...)))..........(((((...)----))))......... ( -22.30, z-score = -2.72, R) >consensus ___UCCAAUGUUAGUAUUUAACAGACAAUUGAUUC______________UUUCACUGGCCGCUUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUCAAG____AGCAUUAUACACU ........................................................((((((.....))))))............................................... (-13.20 = -13.20 + 0.00)

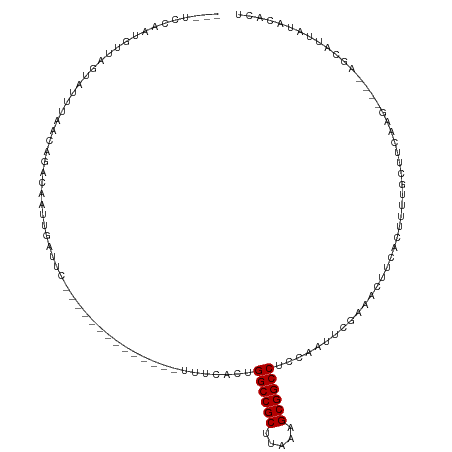
| Location | 10,963,817 – 10,963,929 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.91 |
| Shannon entropy | 0.53251 |
| G+C content | 0.40580 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -17.02 |
| Energy contribution | -16.88 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
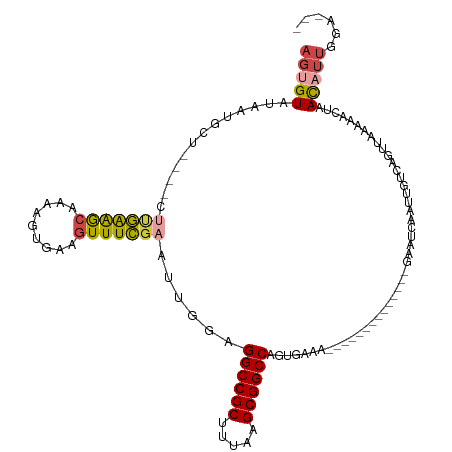
>dm3.chr2L 10963817 112 - 23011544 AGUGUAUAAUGCU----CUUGAGGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCCC-UGAAUUAAUUGUCUGUUAAAAACUAACAUUGGA--- (((((((((((.(----((((((((.........)))))........((((((.....)))))).....))))))))))....-....(((((.....)))))......)))))...--- ( -23.70, z-score = -0.12, R) >droAna3.scaffold_12916 14825746 102 - 16180835 AGUGUAAUGUGGUAAAAAAAAAAAGAAAAGUGCAGUUUUCAAAUUGUGGCCGCUUUACGCGGCCAGAUA----------------UUGUCGCUAGACAG--AAAUACUUAUUGCAGUGGG (.(((((((.((((..........(((((......)))))....(.(((((((.....))))))).)..----------------(((((....)))))--...))))))))))).)... ( -27.80, z-score = -2.16, R) >droEre2.scaffold_4929 12156044 110 + 26641161 AGUGUAUAAUAUU----CGCGAAGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCCCUUGCAUCAACCACCAACAAAAUACUGACAUU------ (((((........----..((((((.........))))))..((((.((((((.....)))))).(..(..(((...)))...)..)........))))....)))))......------ ( -26.10, z-score = -1.37, R) >droYak2.chr2L 7355602 113 - 22324452 AGUGUAUAAGGCU----CUUGAAGCUACAGUGAAGUUUCGAAUUGUAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGCCCCCCUGUAUCAACAACCAACCAAAUACUAACAUUGGA--- ...((((..((..----.((((((((.......)))))))).((((.((((((.....)))))).......(((((.........))))).))))....))..))))..........--- ( -27.30, z-score = -1.01, R) >droSec1.super_3 6364626 99 - 7220098 AGUGUAUAAUGCU----CUUGAAGCAAAAGUGAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAA--------------GAAUCAAUUGUCUGUUAAAAACUAACUUUGGA--- .........((((----.....))))(((((..(((((..((.((((((((((.....))))))..(((..--------------...)))....))))))..))))).)))))...--- ( -26.30, z-score = -1.85, R) >droSim1.chr2L 10760602 99 - 22036055 AGUGUAUAAUGCU----CUUGGAGCAAAAGUGAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAA--------------GAAUCAAUUGUCUGUUAAAAACUAACAUUGGA--- (((((....((((----(...))))).......(((((..((.((((((((((.....))))))..(((..--------------...)))....))))))..))))).)))))...--- ( -27.20, z-score = -2.01, R) >consensus AGUGUAUAAUGCU____CUUGAAGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAAGCGGCCAGUGAAA______________GAAUCAAUUGUCAGUUAAAAACUAACAUUGGA___ (((...............(((((((.........)))))))......((((((.....)))))).........................................)))............ (-17.02 = -16.88 + -0.14)

| Location | 10,963,853 – 10,963,964 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.88 |
| Shannon entropy | 0.45613 |
| G+C content | 0.45739 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
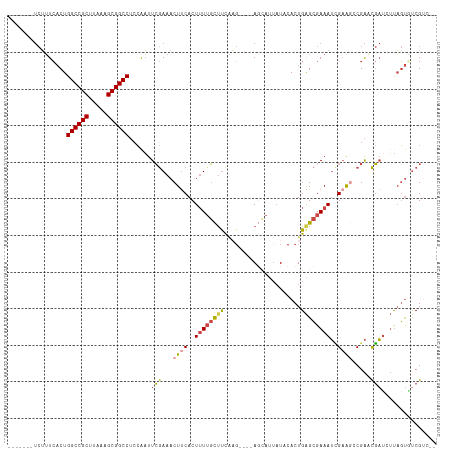
>dm3.chr2L 10963853 111 + 23011544 GACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCCUCAAG----AGCAUUAUACACUGAGCGAAAUCGAAGCCGAACGAUCUUAGUGUCGUC-- ((((.......((...((((((.....)))))).....((((...((((..((((((.(((..----............)))))))))..)))).)))).))......))))...-- ( -29.16, z-score = -2.40, R) >droAna3.scaffold_12916 14825777 108 + 16180835 ---------AAUAUCUGGCCGCGUAAAGCGGCCACAAUUUGAAAACUGCACUUUUCUUUUUUUUUUUACCACAUUACACUGGGAGAAAGGGAGACCGGUUCCUUGCUUCUUCGUCAG ---------......(((((((.....))))))).....(((.....(((((..((((((((((((..(((........)))))))))))))))..))).....)).......))). ( -26.80, z-score = -1.39, R) >droEre2.scaffold_4929 12156078 109 - 26641161 GACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUCGCG----AAUAUUAUACACUGAGCGAAAUUGCAGCCGAUCGAUCUUAGUGUCG---- ((((............((((((.....))))))......((((..((.((.((((((((.(.(----.........).).)))))))).)).))....))))......)))).---- ( -27.70, z-score = -2.10, R) >droYak2.chr2L 7355639 109 + 22324452 GGCAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUACAAUUCGAAACUUCACUGUAGCUUCAAG----AGCCUUAUACACUGAGCGAAAUCGAAGUCGAUCGACCUUAGUGCCG---- (((.............((((((.....))))))........(((.((.......)).)))...----.))).....((((((((((..(((....))))))..)))))))...---- ( -30.70, z-score = -2.35, R) >droSec1.super_3 6364656 104 + 7220098 -------UCUUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUUCAAG----AGCAUUAUACACUGAGCGAAAUCGAAACCGAACGAUGUUAGUAUCGUC-- -------....((...((((((.....))))))..(((((((............(((((...)----))))............)).))))).....))((((((....)))))).-- ( -26.75, z-score = -1.86, R) >droSim1.chr2L 10760632 104 + 22036055 -------UCUUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUCCAAG----AGCAUUAUACACUGAGCGAAAUCGAAGCCGAACGAUGUUAGUAUCGUC-- -------.........((((((.....))))))..((.(((((...(((.(((.(((((...)----)))).........))).))).)))))..)).((((((....)))))).-- ( -29.20, z-score = -2.19, R) >consensus _______UCUUUCACUGGCCGCUUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUCAAG____AGCAUUAUACACUGAGCGAAAUCGAAGCCGAACGAUCUUAGUGUCGUC__ ................((((((.....))))))......(((...((((..((((((((.....................))))))))..)))).....)))............... (-17.08 = -17.50 + 0.42)

| Location | 10,963,853 – 10,963,964 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Shannon entropy | 0.45613 |
| G+C content | 0.45739 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
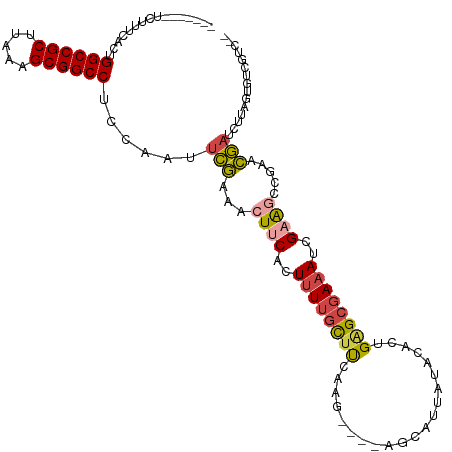
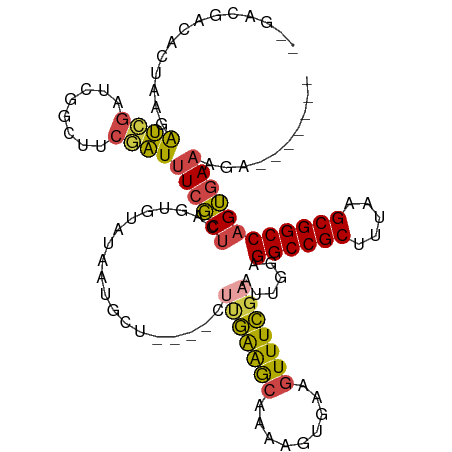
>dm3.chr2L 10963853 111 - 23011544 --GACGACACUAAGAUCGUUCGGCUUCGAUUUCGCUCAGUGUAUAAUGCU----CUUGAGGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUC --(((((..((((..(((...(((((((.(((.((((((.((.....)).----).))).)).))).))))))).))).)))).((((((.....))))))...........))))) ( -33.30, z-score = -1.29, R) >droAna3.scaffold_12916 14825777 108 - 16180835 CUGACGAAGAAGCAAGGAACCGGUCUCCCUUUCUCCCAGUGUAAUGUGGUAAAAAAAAAAAGAAAAGUGCAGUUUUCAAAUUGUGGCCGCUUUACGCGGCCAGAUAUU--------- .....(((((.(((.(((.......))).(((..((((......)).))..))).............)))..)))))....(.(((((((.....))))))).)....--------- ( -24.00, z-score = -0.24, R) >droEre2.scaffold_4929 12156078 109 + 26641161 ----CGACACUAAGAUCGAUCGGCUGCAAUUUCGCUCAGUGUAUAAUAUU----CGCGAAGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUC ----.((((((((..((((..((((.((.(((.(((..(((.........----)))..))).))).)).)))))))).)))).((((((.....))))))............)))) ( -32.20, z-score = -1.60, R) >droYak2.chr2L 7355639 109 - 22324452 ----CGGCACUAAGGUCGAUCGACUUCGAUUUCGCUCAGUGUAUAAGGCU----CUUGAAGCUACAGUGAAGUUUCGAAUUGUAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGCC ----.((((.(((((((....)))))...(((((((...........((.----.((((((((.......))))))))...)).((((((.....)))))))))))))..)).)))) ( -34.80, z-score = -1.75, R) >droSec1.super_3 6364656 104 - 7220098 --GACGAUACUAACAUCGUUCGGUUUCGAUUUCGCUCAGUGUAUAAUGCU----CUUGAAGCAAAAGUGAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAAGA------- --((((((......))))))((((((.(((((((((...(((........----......)))..)))))))))..))))))..((((((.....)))))).........------- ( -31.44, z-score = -1.56, R) >droSim1.chr2L 10760632 104 - 22036055 --GACGAUACUAACAUCGUUCGGCUUCGAUUUCGCUCAGUGUAUAAUGCU----CUUGGAGCAAAAGUGAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAAGA------- --.....((((....(((((((((((((.(((.((((((.((.....)).----))..)))).))).))))))..)))).))).((((((.....)))))))))).....------- ( -33.60, z-score = -1.76, R) >consensus __GACGACACUAAGAUCGAUCGGCUUCGAUUUCGCUCAGUGUAUAAUGCU____CUUGAAGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAAGCGGCCAGUGAAAGA_______ ..............((((........))))((((((...................(((((((.........)))))))......((((((.....)))))))))))).......... (-21.30 = -21.08 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:19 2011