| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,821,595 – 10,821,694 |
| Length | 99 |
| Max. P | 0.974093 |

| Location | 10,821,595 – 10,821,694 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.31 |
| Shannon entropy | 0.67086 |
| G+C content | 0.48616 |
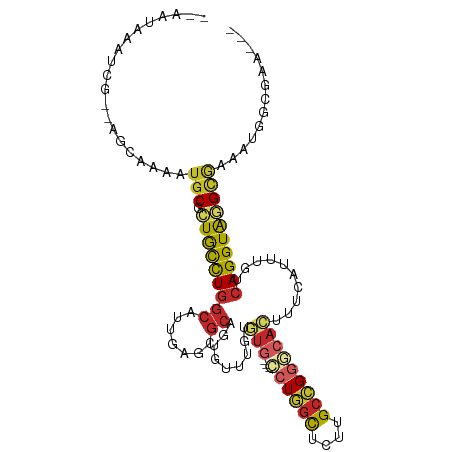
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.51 |
| Covariance contribution | 0.22 |
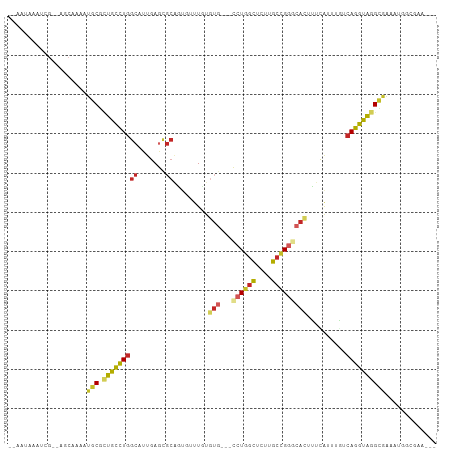
| Combinations/Pair | 1.52 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
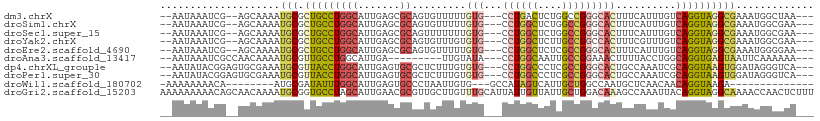
>dm3.chrX 10821595 99 + 22422827 --AAUAAAUCG--AGCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUG---CCUGACUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCUAA--- --.........--(((....(((.((((((((((.((((...((((((..((.(---((.......)))))))))))))))..))))))))))))).....)))..--- ( -34.60, z-score = -1.31, R) >droSim1.chrX 8384714 99 + 17042790 --AAUAAAUCG--AGCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUG---CCUGGCUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCGAA--- --......(((--..((...(((.((((((((((.(((((.....))....(((---((((((....)))))))))..)))..)))))))))))))...)).))).--- ( -41.00, z-score = -2.56, R) >droSec1.super_15 1524590 99 + 1954846 --AAUAAAUCG--AGCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUUUUUUUGUG---CCUGGCUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCGAA--- --......(((--..((...(((.((((((((((.((((............(((---((((((....))))))))).))))..)))))))))))))...)).))).--- ( -40.82, z-score = -2.69, R) >droYak2.chrX 10471401 99 - 21770863 --AAUAAAUCG--AGCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUGUGUG---CCUGGCUCUUGCCGGCCACUUUCGUUUGUCAGGUAGGCGAAAUGGCGAA--- --......(((--..((...(((.((((((((((.(((((((((...))))))(---((.(((....)))))).....)))..)))))))))))))...)).))).--- ( -36.40, z-score = -1.13, R) >droEre2.scaffold_4690 14441657 99 - 18748788 --AAUAAAUCG--AGCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUG---CCUGGCUCUCGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGGGAA--- --......(((--.((.....)).((((((((((.(((((.....))....(((---((((((....)))))))))..)))..)))))))))).))).........--- ( -38.10, z-score = -1.93, R) >droAna3.scaffold_13417 2351113 92 + 6960332 --AAUAAAUCGCCAACAAAAUGCGUUGCCUGGCAUUGA---------UUGUAUA---CCUGGCAAUUGCCGGAAACUUUUACCUGGCAGGUGAGUAAUUCAAAAAA--- --......(((((..........((((((.((.((...---------....)).---)).))))))((((((..........))))))))))).............--- ( -24.10, z-score = -1.44, R) >dp4.chrXL_group1e 648028 101 + 12523060 --AAUAUACGGAGUGCGAAAUGCGUUACCUGGCAUUGAGUGCGCUCUUUGUGUG---CCUGGCCCUCGCCGGGCACUGCCAAAUCGCAGGUAAGUGGAUAGGGUCA--- --...(((.((((((((.(((((........)))))...)))))))).)))(((---((((((....))))))))).(((...((((......))))....)))..--- ( -40.50, z-score = -1.75, R) >droPer1.super_30 388928 101 - 958394 --AAUAUACGGAGUGCGAAAUGCGUUACCUGGCAUUGAGUGCGCUCUUUGUGUG---CCUGGCCCUCGCCGGGCACUGCCAAAUCGCAGGUAAGUGGAUAGGGUCA--- --...(((.((((((((.(((((........)))))...)))))))).)))(((---((((((....))))))))).(((...((((......))))....)))..--- ( -40.50, z-score = -1.75, R) >droWil1.scaffold_180702 874450 83 + 4511350 -AAAAAAAACA--------AUGCGAUAUUUGGCAUUGAGUGCCCUAAUUGUG---GCCAUAGUCAUUGCUGGCCAAUGCUCAACAACAGGUAAGA-------------- -........((--------((((.(....).))))))..((((.......((---((((..........)))))).((.....))...))))...-------------- ( -18.10, z-score = 0.22, R) >droGri2.scaffold_15203 6850468 109 + 11997470 AAAAAAAAACAGCAACAAAAUGCGGUGCCUAGCAUUGAACGCGUUGCUUGUUUGCAUUAUUGUUAUUGCUGGACAAAGCCAAAUUACAGGUAGGCAAAACCAACUCUUU .........((((((((((((((((.((..((((.((....)).)))).))))))))).))))...)))))......(((............))).............. ( -22.50, z-score = 0.67, R) >consensus __AAUAAAUCG__AGCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUGUGUG___CCUGGCUCUUGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCGAA___ ....................(((.(((((((((.......)).........(((...((((((....)))))))))..........))))))))))............. (-17.29 = -17.51 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:37 2011