| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,810,845 – 10,810,950 |
| Length | 105 |
| Max. P | 0.939769 |

| Location | 10,810,845 – 10,810,950 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Shannon entropy | 0.56621 |
| G+C content | 0.61448 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.81 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
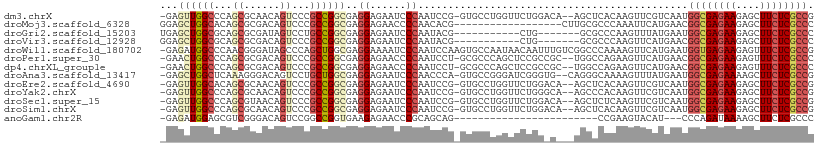
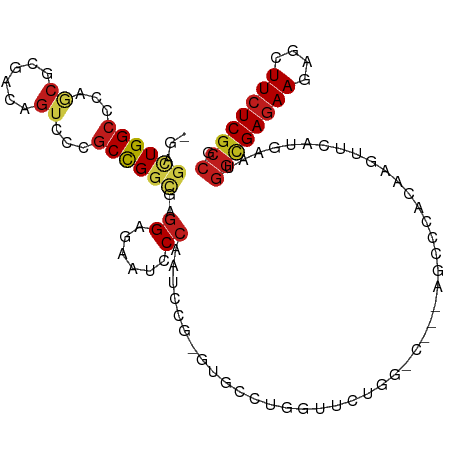
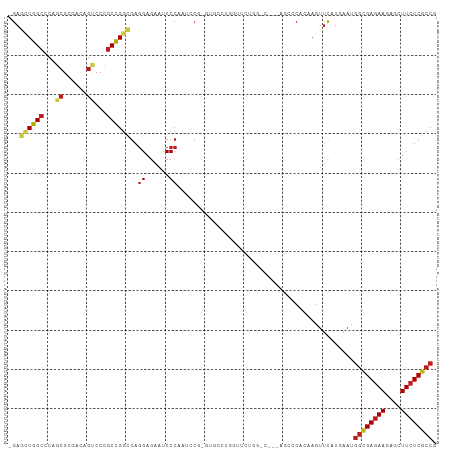
>dm3.chrX 10810845 105 + 22422827 -GAGUUGGCCCAGCGCAACAGUCCCGCCGGCGAGGAGAAUCCCAAUCCG-GUGCCUGGUUCUGGACA--AGCUCACAAGUUCGUCAAUGGCGAGAAGAGCUUCUCGCCG -(((((.(.((((.((..(((...((((((...((......))...)))-))).))))).)))).).--)))))..............((((((((....)))))))). ( -39.90, z-score = -1.52, R) >droMoj3.scaffold_6328 4007314 91 - 4453435 GGAGCUGGCACAGCGCGACAGUCCCGCCGGCGAGGAGAACCCCAACACG------------------CUUGCGCCCAAAUUCAUGAACGGCGAGAAGAGCUUCUCGCCC ((..(((((.....))..)))..))((.((((.((......))....))------------------)).))................((((((((....)))))))). ( -29.60, z-score = -0.31, R) >droGri2.scaffold_15203 6838465 91 + 11997470 UGAGCUGGCGCAGCGCGAUAGUCCUGCCGGCGAGGAGAAUCCCAAUACG-----------CUG-------GCGCCCAAGUUUAUGAAUGGCGAGAAGAGCUUCUCGCCC ((((((((.((((.((....)).))(((((((.((......))....))-----------)))-------)))))).)))))).....((((((((....)))))))). ( -37.20, z-score = -2.05, R) >droVir3.scaffold_12928 406334 91 + 7717345 GGAGCUGGCGCAGCGCGACAGUCCCGCCGGCGAGGAGAAUCCCAAUACG-----------CUG-------GCGCCCAAGUUCAUGAACGGCGAGAAGAGCUUCUCGCCC ((..(((.(((...))).)))..))(((((((.((......))....))-----------)))-------))......(((....)))((((((((....)))))))). ( -39.20, z-score = -1.82, R) >droWil1.scaffold_180702 862669 108 + 4511350 -GAGAUGGCCCAACGGGAUAGCCCAGCUGGCGAGGAAAAUCCCAAUCCAAGUGCCAAUAACAAUUUGUCGGCCCAAAAGUUCAUGAAUGGUGAGAAGAGUUUCUCGCCG -(((.(((.((.(((((....)))...(((((.(((.........)))...)))))..........)).)).)))....)))......((((((((....)))))))). ( -30.70, z-score = -1.19, R) >droPer1.super_30 377699 105 - 958394 -GAACUGGCCCAGCGCGACAGUCCCGCCGGCGAGGAGAACCCCAAUCCU-GCGCCCAGCUCCGCCGC--UGGCCAGAAGUUCAUGAACGGCGAGAAGAGUUUCUCGCCC -...((((((.((((((.......))).((((((((.........))))-((.....))..))))))--)))))))..(((....)))((((((((....)))))))). ( -44.50, z-score = -2.97, R) >dp4.chrXL_group1e 636740 105 + 12523060 -GAACUGGCCCAGCGCGACAGUCCCGCCGGCGAGGAGAACCCCAAUCCU-GCGCCCAGCUCCGCCGC--UGGCCAGAAGUUCAUGAACGGCGAGAAGAGUUUCUCGCCC -...((((((.((((((.......))).((((((((.........))))-((.....))..))))))--)))))))..(((....)))((((((((....)))))))). ( -44.50, z-score = -2.97, R) >droAna3.scaffold_13417 2339872 105 + 6960332 -GAGCUGGCUCAAAGGGACAGUCCUGCUGGCGAGGAGAAUCCCAACCCA-GUGCCGGGAUCGGGUG--CAGGGCAAAAGUUUAUGAAUGGCGAGAAAAGCUUCUCGCCG -(((((.((((...((((...(((((....).))))...))))......-(..((.......))..--).))))...)))))......((((((((....)))))))). ( -39.50, z-score = -1.13, R) >droEre2.scaffold_4690 14432055 105 - 18748788 -GAGUUGGCACAGCGCAACAGUCCCGCCGGCGAGGAGAAUCCCAAUCCG-GUGCCUGGUUCUGGACA--AGCUCACAAGUUCGUCAAUGGCGAGAAGAGCUUCUCGCCG -(((((.((.....))....((((.(((((((.(((.........))).-.)))).)))...)))).--)))))..............((((((((....)))))))). ( -38.50, z-score = -1.18, R) >droYak2.chrX 10462087 105 - 21770863 -GAGUUGGCCCAGCGCAACAGUCCCGCCGGCGAGGAGAAUCCCAAUCCG-GUGCCUGGUUCUGGGCA--AGCCCACAAGUUCGUCAAUGGCGAGAAGAGCUUCUCGCCG -(.(((.((((((.((..(((...((((((...((......))...)))-))).))))).)))))).--))).)..............((((((((....)))))))). ( -42.20, z-score = -1.74, R) >droSec1.super_15 1514158 105 + 1954846 -GAGUUGGCCCAGCGUAACAGUCCCGCCGGCGAGGAGAAUCCCAAUCCG-GUGCCUGGUUCUGGACA--AGCUCUCAAGUUCGUCAAUGGCGAGAAGAGCUUCUCGCCG -(((((.(.((((.....(((...((((((...((......))...)))-))).)))...)))).).--)))))..............((((((((....)))))))). ( -38.60, z-score = -1.30, R) >droSim1.chrX 8373299 105 + 17042790 -GAGUUGGCCCAGCGCAACAGUCCCGCCGGCGAGGAGAAUCCCAAUCCG-GUGCCUGGUUCUGGACA--AGCUCACAAGUUCGUCAAUGGCGAGAAGAGCUUCUCGCCG -(((((.(.((((.((..(((...((((((...((......))...)))-))).))))).)))).).--)))))..............((((((((....)))))))). ( -39.90, z-score = -1.52, R) >anoGam1.chr2R 9514844 81 - 62725911 -GAGAUGGAGCGUCGGGACAGUCCGGCCGGUGAAGAGAACCCGCAGCAG------------------------CCGAAGUACAU---CCCAGAUAAAAGCUUCUCGCCC -..((.((((((((((((.....((((.(.((..(.....)..)).).)------------------------))).......)---))).)))....))))))).... ( -20.80, z-score = 0.35, R) >consensus _GAGCUGGCCCAGCGCGACAGUCCCGCCGGCGAGGAGAAUCCCAAUCCG_GUGCCUGGUUCUGG_C___AGCCCACAAGUUCAUGAAUGGCGAGAAGAGCUUCUCGCCG ...((((((...((......))...))))))..((......)).............................................((((((((....)))))))). (-20.30 = -20.81 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:35 2011