| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,752,461 – 10,752,584 |
| Length | 123 |
| Max. P | 0.901269 |

| Location | 10,752,461 – 10,752,584 |
|---|---|
| Length | 123 |
| Sequences | 7 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 72.38 |
| Shannon entropy | 0.52663 |
| G+C content | 0.57092 |
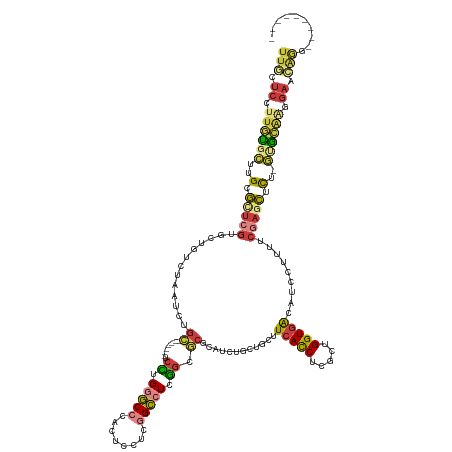
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.50 |
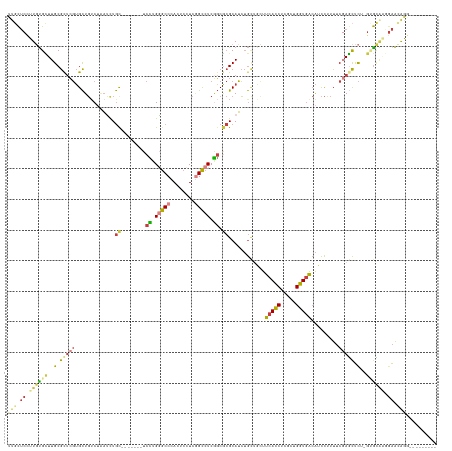
| Combinations/Pair | 1.44 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
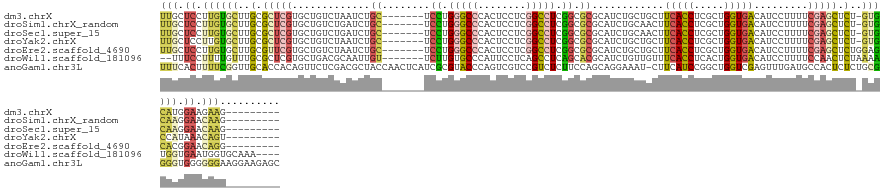
>dm3.chrX 10752461 123 - 22422827 UUGCUCCUUGUGCUUGCGCUCGUGCUGUCUAAUCUGC-------UCCUGGGCCCACUCCUCGGCCUCGGCGCGCAUCUGCUGCUUCACCUCGCUGGUGACAUCCUUUUCGAGCUCU-GUGCAUGGAAGAAG--------- ....(((.((..(..(.(((((.((..........((-------.((.(((((........))))).)).))((....)).)).(((((.....))))).........))))).).-)..)).))).....--------- ( -40.80, z-score = -1.27, R) >droSim1.chrX_random 3030570 123 - 5698898 UUGCUCCUUGUGCUUGCGCUCGUGCUGUCUGAUCUGC-------UCCUGGGCCCACUCCUCGGCCUCGGCGCGCAUCUGCAACUUCACCUCGCUGGUGACAUCCUUUUCGAGCUCU-GUGCAAGGAACAAG--------- (((.((((((..(..(.((((((((.....(((.(((-------.((.(((((........))))).)).))).))).)))...(((((.....))))).........))))).).-)..)))))).))).--------- ( -49.30, z-score = -4.08, R) >droSec1.super_15 1455745 123 - 1954846 UUGCUCCUUGUGCUUGCGCUCGUGCUGUCUGAUCUGC-------UCCUGGGCCCACUCCUCGGCCUCGGCGCGCAUCUGCAACUUCACCUCGCUGGUGACAUCCUUUUCGAGCUCU-GUGCAAGGAACAAG--------- (((.((((((..(..(.((((((((.....(((.(((-------.((.(((((........))))).)).))).))).)))...(((((.....))))).........))))).).-)..)))))).))).--------- ( -49.30, z-score = -4.08, R) >droYak2.chrX 19324222 123 - 21770863 UUGCUCCUUGUGCUUGCGCUCGUGCUGUCUAAUCUGC-------UCCUGGGCCCACUCCUCGGCCUCGGCGCGCAUCUGCUGCUUCACCUCGCUGGUGACAUCCUUUUCGAGCUCU-GUGCCAUAAACAGU--------- ..((((...((((.(((((....))..........((-------.((.(((((........))))).)).)))))...)).)).(((((.....)))))..........)))).((-((.......)))).--------- ( -36.40, z-score = -1.05, R) >droEre2.scaffold_4690 14372038 124 + 18748788 UUGCUCCUUGUGCUUGCGUUCGUGCUGUCUAAUCUGC-------UCCUGGGCCCACUCCUCGGCCUCGGCGCGCAUCUGCUGCUUCACCUCGCUGGUGACAUCCUUUUCGAGCUCUGGAGCACGGAACAGG--------- .((((((..(.((((((((..((((.(......).((-------.((.(((((........))))).)).))))))..)).)).(((((.....)))))..........)))).).)))))).........--------- ( -41.50, z-score = -0.76, R) >droWil1.scaffold_181096 4941791 127 + 12416693 --UUUCCUUUUGUUUGCGCUCGUGCUGACGCAAUUGU-------UCUUGUGCCCAUUCCUCAGCCUCAGCACGCAUCUGUUGUUUCACCUCACUGGUGACAUCCUUUUCCAACUCUAAAAUGGUGAAUGGUGCAAA---- --..........((((((((((((((((.((......-------..................)).))))))))...........(((((.....)))))((((.((((........)))).))))...))))))))---- ( -30.56, z-score = -1.38, R) >anoGam1.chr3L 19607547 139 - 41284009 UUUCACUUUUCGGUUGCACCACAGUUCUCGACGCUACCAACUCAUCGCGUACCCAGUCGUCCGUCUCUUCCAGCAGGAAAU-CUUCAUCCGGCUGGUCGAGUUUGAUGCCACUCUCUGCGGGGUGGGGGGAAGGAAGAGC ((((.(((..((((.((...............)).)))..((((((.((((...(((((((...(((..(((((.(((...-.....))).)))))..)))...))))..)))...)))).)))))))..)))))))... ( -46.16, z-score = -0.54, R) >consensus UUGCUCCUUGUGCUUGCGCUCGUGCUGUCUAAUCUGC_______UCCUGGGCCCACUCCUCGGCCUCGGCGCGCAUCUGCUGCUUCACCUCGCUGGUGACAUCCUUUUCGAGCUCU_GUGCAAGGAACAGG_________ .(((.......(((((....(((((((.....................(((((........))))))))))))...........(((((.....))))).........)))))......))).................. (-19.40 = -19.90 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:27 2011