
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,747,390 – 10,747,448 |
| Length | 58 |
| Max. P | 0.711211 |

| Location | 10,747,390 – 10,747,448 |
|---|---|
| Length | 58 |
| Sequences | 10 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Shannon entropy | 0.59473 |
| G+C content | 0.54502 |
| Mean single sequence MFE | -15.02 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.21 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10747390 58 + 22422827 GCCUCGUAUGCCUGCACCAGUUCCGUGAUCUGCUGCUCAAUCUUGGGCAGCUUGGAGG .((((.........(((.......)))....((((((((....))))))))...)))) ( -18.30, z-score = -0.53, R) >droSec1.super_15 1450693 58 + 1954846 GCCUCGUAUGCCUGAACCAGUUCCGUGAUCUGCUGCUCAAUCUUGGGCAGCUUGGAGG .((((.((((...(((....)))))))....((((((((....))))))))...)))) ( -18.00, z-score = -0.84, R) >droYak2.chrX 19319156 58 + 21770863 GCCUCGUAUGCCUGCACCAGCUCCGUGAUCUGCUGCUCAAUCUUGGGCAGCUUGGAGG .((((.((((...((....))..))))....((((((((....))))))))...)))) ( -19.70, z-score = -0.74, R) >droEre2.scaffold_4690 14367017 58 - 18748788 ACCUCGUAUGCCUGUACCAGCUCCGUGAUCUGCUGCUCAAUCUUGGGCAGCUUGGAGG ..((.((((....)))).))(((((......((((((((....)))))))).))))). ( -19.80, z-score = -1.55, R) >droAna3.scaffold_13417 1358622 58 + 6960332 GCCUCGAAAGCCUGCACCAGCUCCGUGAUCUGCUGCUCGAUCUUGGGCAGCUUCGACG ...((((((((........))).........((((((((....))))))))))))).. ( -16.70, z-score = -0.27, R) >droPer1.super_11 447856 58 + 2846995 ACCUCAUAGGCGUGCACCAGUUCAGUUAUCUGCUGCUCAGUCUUGGGCAGCUUCGAGG .((((...((......)).............(((((((......)))))))...)))) ( -18.80, z-score = -1.35, R) >droWil1.scaffold_180777 4350904 58 + 4753960 UCCUGAUAUUCCUGAACAAGCUCGGUUAUUUCCUGUUCAAUUUUGGGCAGCUUCGAGG .(((((.((..((((......))))..)).))(((((((....))))))).....))) ( -11.50, z-score = -0.12, R) >droVir3.scaffold_12970 10353207 58 + 11907090 AUCUCAUAGGCUUGCACCAAUUCGGUAAUUUGCUGCUCUAUCUUUGGCAGCUUAGUGC .............(((((.....))).....((((((........)))))).....)) ( -11.10, z-score = 0.38, R) >droMoj3.scaffold_6473 5836736 58 - 16943266 AUCUCAUACGCUUUCACCAACUCUGUGAUCUGUUGCUCAAUCUUUGGCAAUUUGGUCC ..........................((((.((((((........))))))..)))). ( -8.40, z-score = -0.80, R) >anoGam1.chrU 52785973 53 + 59568033 -CUCCCGCUGCAUCCACU-GCUCUUCGAUCAGCUCCUCC-UCCGGCGUACCGUACA-- -....(((((........-(((........)))......-..))))).........-- ( -7.89, z-score = -0.49, R) >consensus ACCUCGUAUGCCUGCACCAGCUCCGUGAUCUGCUGCUCAAUCUUGGGCAGCUUGGAGG ...............................((((((((....))))))))....... ( -9.07 = -9.21 + 0.14)

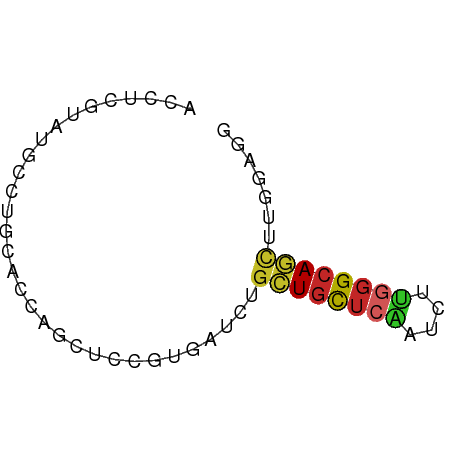
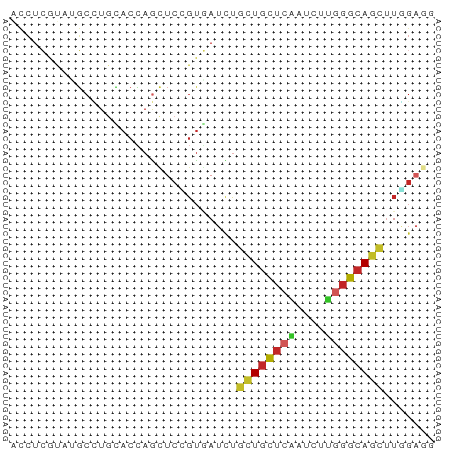
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:26 2011