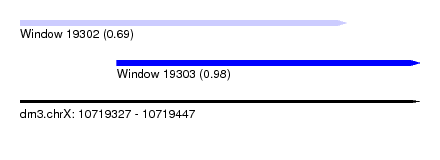
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,719,327 – 10,719,447 |
| Length | 120 |
| Max. P | 0.979576 |

| Location | 10,719,327 – 10,719,425 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.47258 |
| G+C content | 0.57419 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -12.90 |
| Energy contribution | -14.03 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10719327 98 + 22422827 --AUGGAAAUG--GGAAAGGA--AGAGCCACCACCCACCCCCA-GCGGCAACUGCGACGCAUUUUUGUGUGUGCGUCCUGUGCGUU-GCCAUGCUGCGAUUUUCCA --.((((((((--((...((.--.(......)..))...))))-((((((...(((((((((...(((....)))....)))))))-))..))))))...)))))) ( -35.90, z-score = -1.83, R) >droAna3.scaffold_13417 1322623 98 + 6960332 ACGCGGCCGGGAGGUCCUUGUUUAAAGCCACCACCCACCCCCGGGUGGCAACUGCGACGCAUUUUUCUGCGU-UGCGUUUUGCGUUUGACAGGCUGCGA------- .((((((((((....)))((((....((..((((((......)))))).....((((((((......)))))-))).....))....))))))))))).------- ( -45.20, z-score = -3.12, R) >droEre2.scaffold_4690 14337915 99 - 18748788 -AAGGGAAGGGCAGGGAAGGG--AAAGCCACCACCCACCCCU--GAGGCAACUGCGACGCAUUUUUGUGUGUGCGUCCUGUGCGUU-GCCAUGCUGCGAUUUCCA- -..(((((..((((((..(((--..........)))..))))--..((((((.(((((((((........)))))))....)))))-))).....))..))))).- ( -39.50, z-score = -1.20, R) >droSec1.super_15 1422760 84 + 1954846 --AUGGAAAUG--GGAAAGGG--AAAGCCACCACCCACCCCU--GCGGCAACUGCGACGCAUUUUUG-------------UGCGUU-GCCAUGCUGCGAUUUUCCA --.((((((((--((....((--....))....))))....(--((((((...(((((((((....)-------------))))))-))..)))))))..)))))) ( -35.90, z-score = -3.65, R) >consensus __AUGGAAAGG__GGAAAGGG__AAAGCCACCACCCACCCCC__GCGGCAACUGCGACGCAUUUUUGUGUGU_CGUCCUGUGCGUU_GCCAUGCUGCGAUUUUCC_ ............................................((((((...(((((((((.................))))))).))..))))))......... (-12.90 = -14.03 + 1.13)

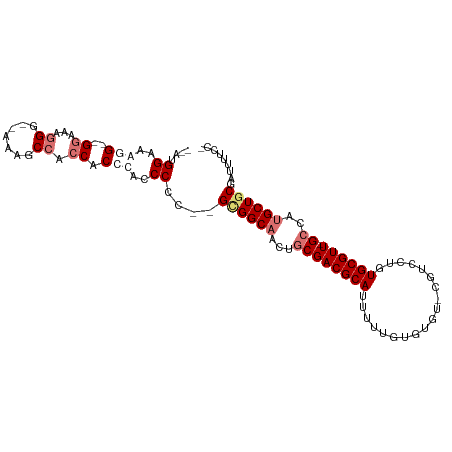
| Location | 10,719,356 – 10,719,447 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.24 |
| Shannon entropy | 0.44361 |
| G+C content | 0.54159 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -18.86 |
| Energy contribution | -21.30 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
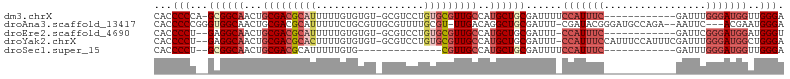
>dm3.chrX 10719356 91 + 22422827 CACCCCCA-GCGGCAACUGCGACGCAUUUUUGUGUGU-GCGUCCUGUGCGUUGCCAUGCUGCGAUUUUCCAUUUC------------GAUUUGGGAUGGUUGGGA ....((((-((((((...(((((((((...(((....-)))....)))))))))..))))))......((((..(------------.....)..)))).)))). ( -37.40, z-score = -2.72, R) >droAna3.scaffold_13417 1322658 98 + 6960332 CACCCCCGGGUGGCAACUGCGACGCAUUUUUCUGCGUUGCGUUUUGCGU-UUGACAGGCUGCGAUUU-CGAUACGGGAUGCCAGA--AAUUC---ACGAAUGGGA ....((((..((((((.(((((((((......)))))))))..))))..-.(((....(((..((..-((...))..))..))).--...))---)))..)))). ( -31.10, z-score = -0.37, R) >droEre2.scaffold_4690 14337947 89 - 18748788 CACCCCU--GAGGCAACUGCGACGCAUUUUUGUGUGU-GCGUCCUGUGCGUUGCCAUGCUGCGAUUU-CCAUUUC------------GAUUCGGGAUGGAUGGGU .((((..--..((((((.(((((((((........))-)))))....))))))))...........(-((((..(------------(...))..))))).)))) ( -35.50, z-score = -2.39, R) >droYak2.chrX 19291068 101 + 21770863 CACCCCU--GAGGCAACUGCGACGCACUUUUGUGUGU-GCGUCCUGUGCGUUGCCAUGCUGCGAUUU-CCAUUUCCAUUUCCAUUUCGAUUUGGGAUGGCUGGGA ...(((.--..((((((.(((((((((........))-)))))....))))))))............-(((((.(((..((......))..))))))))..))). ( -36.60, z-score = -2.31, R) >droSec1.super_15 1422789 77 + 1954846 CACCCCU--GCGGCAACUGCGACGCAUUUUUGUG--------------CGUUGCCAUGCUGCGAUUUUCCAUUUC------------GAUUUGGGAUGGUUGGGA ...((((--((((((...(((((((((....)))--------------))))))..))))))).....((((..(------------.....)..))))..))). ( -34.80, z-score = -4.33, R) >consensus CACCCCU__GAGGCAACUGCGACGCAUUUUUGUGUGU_GCGUCCUGUGCGUUGCCAUGCUGCGAUUU_CCAUUUC____________GAUUUGGGAUGGAUGGGA ...(((...((((((...(((((((((..................)))))))))..))))))......((((((...................))))))..))). (-18.86 = -21.30 + 2.44)
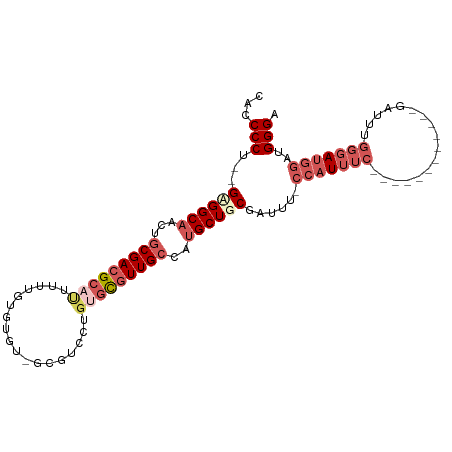
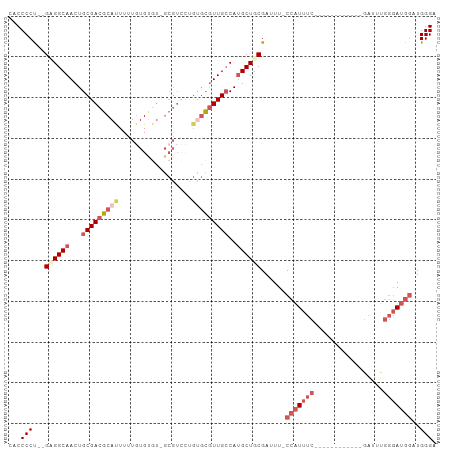
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:24 2011