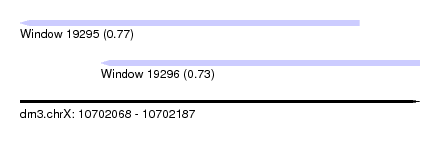
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,702,068 – 10,702,187 |
| Length | 119 |
| Max. P | 0.771079 |

| Location | 10,702,068 – 10,702,169 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 62.46 |
| Shannon entropy | 0.65299 |
| G+C content | 0.56164 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -14.10 |
| Energy contribution | -15.18 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10702068 101 - 22422827 --AUCCCCGUGAUCGUGGCGUGU-UCUUCGACACGCAGGUA-GGACCUCGAAAUUAGCUGAUAGGGUCCUGUUCCCGAUCUCGAUCCCGAUCCGAUCCUGUCCUC-------------- --.....((.(((((.(((((((-(....))))))).((((-((((((...............))))))))...))...).))))).))................-------------- ( -29.66, z-score = -0.14, R) >droSim1.chrX 8320767 101 - 17042790 --AUCCCCGUGAUCGUGGCGUGU-UCUUCGACACGCAGGUA-GGACCUCGAAAUUAGCUGAUAGGGUCCUGUUCCCGAUCUCGAUCCCGAUCCGAUCCUGUCCUC-------------- --.....((.(((((.(((((((-(....))))))).((((-((((((...............))))))))...))...).))))).))................-------------- ( -29.66, z-score = -0.14, R) >droYak2.chrX 19272456 115 - 21770863 --AUCCCCGUGAUCGUGGCGUGU-UCUUCGACACGCAGGUA-GGACCUCGAAAUUGGCUGAUAGGGUCCUGUUCCUGAUCUCGAUCUCGAUCCCGAUCCGCCAUCCCGAUCCUGUCCUC --...............((((((-(....)))))))(((((-(((..(((....((((.(((.(((((..(.((........)).)..)).))).))).))))...)))))))).))). ( -35.20, z-score = -0.22, R) >droAna3.scaffold_13417 1298821 115 - 6960332 UGCUCACUGCGAUCCUUGCGUGU-UC--CGACACGCAGGUA-GGGCCCCGAAAUUACCUGAUAGCGUCUCGUACUCUGUCCUGUGCCCUAUCCCUAUCCCUAUCGCUAUCCCUGUCCCG ........(((((...(((((((-..--..)))))))((((-((((.........((..(((((.((.....)).)))))..)))))))))).........)))))............. ( -31.80, z-score = -3.04, R) >dp4.chrXL_group1e 4919806 79 + 12523060 -----------GUCCUUGCGUGUGUUCUCGACACGCAGGUAGGGGCAUAGAAAUUAAGUGA------CCCGCAAC--AACAAGGACUCGACUCUCUCC--------------------- -----------(((((((.(((((((...)))))))..((.(((((((.........))).------))).).))--..)))))))............--------------------- ( -24.10, z-score = -1.39, R) >consensus __AUCCCCGUGAUCGUGGCGUGU_UCUUCGACACGCAGGUA_GGACCUCGAAAUUAGCUGAUAGGGUCCUGUUCCCGAUCUCGAUCCCGAUCCCAUCCCGUCCUC______________ ..........(((((..((((((.......))))))......((((((...............))))))..................)))))........................... (-14.10 = -15.18 + 1.08)

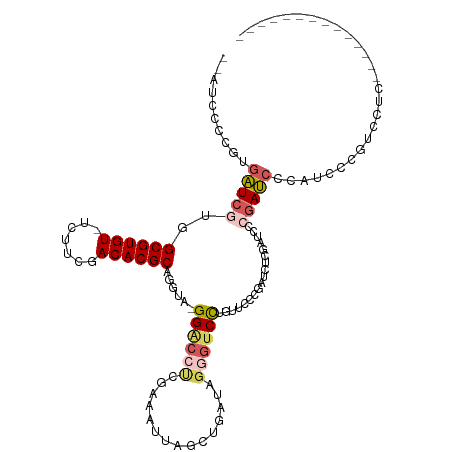

| Location | 10,702,092 – 10,702,187 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.35 |
| Shannon entropy | 0.32572 |
| G+C content | 0.55793 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10702092 95 - 22422827 ----------------------UUCCCGCUGAUCUUCGA--GAUCCCCGUGAUCGUGGCGUGUUCUUCGACACGCAGGUAGGACCUCGAAAUUAGCUGAUAGGGUCCUGUUCCCGAUCU ----------------------.....((((((.(((((--(.(((.....(((...(((((((....))))))).))).))).)))))))))))).(((.(((.......))).))). ( -35.00, z-score = -2.27, R) >droSim1.chrX 8320791 95 - 17042790 ----------------------UUCCCGCUGAUCUUCGA--GAUCCCCGUGAUCGUGGCGUGUUCUUCGACACGCAGGUAGGACCUCGAAAUUAGCUGAUAGGGUCCUGUUCCCGAUCU ----------------------.....((((((.(((((--(.(((.....(((...(((((((....))))))).))).))).)))))))))))).(((.(((.......))).))). ( -35.00, z-score = -2.27, R) >droYak2.chrX 19272494 95 - 21770863 ----------------------UUCCCGCUGAUCUUUAA--GAUCCCCGUGAUCGUGGCGUGUUCUUCGACACGCAGGUAGGACCUCGAAAUUGGCUGAUAGGGUCCUGUUCCUGAUCU ----------------------....((..((((.....--))))..)).((((...(((((((....)))))))(((((((((((...............))))))))..))))))). ( -30.36, z-score = -1.18, R) >droEre2.scaffold_4690 14320605 94 + 18748788 ----------------------UUCCCGCUGAUCUUCAA--GAUCCCCGUGAUCGUGGCGUGUUCUUCGACACGCAGGUAGGACCUCGAAAUUGGCUGAUAGGGUCCUG-UCCCGAUCU ----------------------....((..((((.....--))))..)).(((((..(((((((....))))))).((((((((((...............))))))))-.))))))). ( -32.36, z-score = -1.39, R) >droAna3.scaffold_13417 1298859 117 - 6960332 CCGCCGCCGAGUGUCCGUGGUGUCCCCUAUGAUCCCCGAGUGCUCACUGCGAUCCUUGCGUGUUC--CGACACGCAGGUAGGGCCCCGAAAUUACCUGAUAGCGUCUCGUACUCUGUCC .((..((((((((..((.((..((......))..))))..))))).........(((((((((..--..)))))))))...)))..)).........(((((.((.....)).))))). ( -36.30, z-score = -1.10, R) >consensus ______________________UUCCCGCUGAUCUUCGA__GAUCCCCGUGAUCGUGGCGUGUUCUUCGACACGCAGGUAGGACCUCGAAAUUAGCUGAUAGGGUCCUGUUCCCGAUCU ..............................((((.......)))).....(((((..(((((((....))))))).((((((((((...............))))))))..))))))). (-23.52 = -23.60 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:18 2011