| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,693,854 – 10,693,963 |
| Length | 109 |
| Max. P | 0.695916 |

| Location | 10,693,854 – 10,693,963 |
|---|---|
| Length | 109 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Shannon entropy | 0.47346 |
| G+C content | 0.45483 |
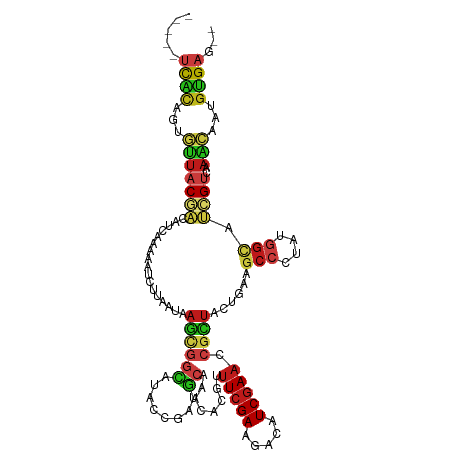
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -11.19 |
| Energy contribution | -11.81 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
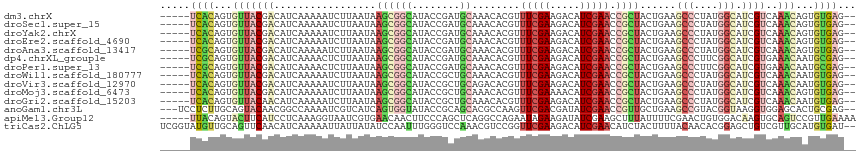
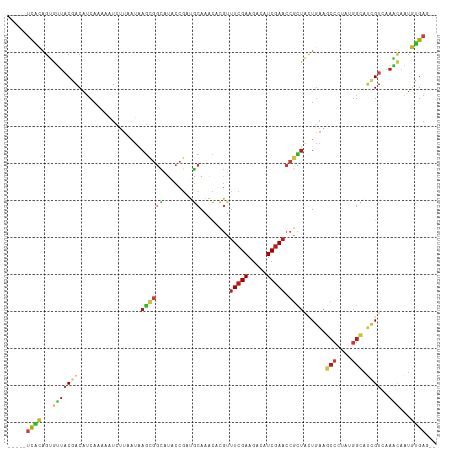
>dm3.chrX 10693854 109 - 22422827 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAGUGUGAG-- -----(((((.((((..(((................((((((((....))))........(((((.....))))).))))...((.(((....))).))))).)))).))))).-- ( -30.30, z-score = -2.94, R) >droSec1.super_15 1391257 109 - 1954846 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAGUGUGAG-- -----(((((.((((..(((................((((((((....))))........(((((.....))))).))))...((.(((....))).))))).)))).))))).-- ( -30.30, z-score = -2.94, R) >droYak2.chrX 19263622 109 - 21770863 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAGUGUGAG-- -----(((((.((((..(((................((((((((....))))........(((((.....))))).))))...((.(((....))).))))).)))).))))).-- ( -30.30, z-score = -2.94, R) >droEre2.scaffold_4690 14312104 109 + 18748788 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAGUGUGAG-- -----(((((.((((..(((................((((((((....))))........(((((.....))))).))))...((.(((....))).))))).)))).))))).-- ( -30.30, z-score = -2.94, R) >droAna3.scaffold_13417 1288984 109 - 6960332 -----UCGCAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAGUGUGAG-- -----(((((.((((..(((................((((((((....))))........(((((.....))))).))))...((.(((....))).))))).)))).))))).-- ( -29.90, z-score = -2.36, R) >dp4.chrXL_group1e 4908801 109 + 12523060 -----UCGCAGUGUUACGACAUCAAAACUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUUCGGCAUCGUGAAACAAUGCGAG-- -----(((((.((((((((.................((((((((....))))........(((((.....))))).))))......(((....))).)))))..))).))))).-- ( -28.70, z-score = -1.90, R) >droPer1.super_13 967571 109 - 2293547 -----UCGCAGUGUUACGACAUCAAAACUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUUCGGCAUCGUGAAACAAUGCGAG-- -----(((((.((((((((.................((((((((....))))........(((((.....))))).))))......(((....))).)))))..))).))))).-- ( -28.70, z-score = -1.90, R) >droWil1.scaffold_180777 3034611 109 - 4753960 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGCUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAAUGUGAG-- -----(((((.((((..(((.(((.............(((((.....))))).....((.(((((.....))))).))....))).(((....)))...))).)))).))))).-- ( -29.40, z-score = -2.98, R) >droVir3.scaffold_12970 10286669 109 - 11907090 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGCUGCAGACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAAUGUGAG-- -----(((((.((((......................(((((.....))))).(((.((.(((((.....))))).)).....((.(((....))).))))).)))).))))).-- ( -30.40, z-score = -2.91, R) >droMoj3.scaffold_6473 5772183 109 + 16943266 -----UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGCUGCAAACACGUUUCGAAAACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAGUGUGAG-- -----(((((.((((..(((.(((.............(((((.....))))).....((.(((((.....))))).))....))).(((....)))...))).)))).))))).-- ( -31.50, z-score = -3.59, R) >droGri2.scaffold_15203 6580878 109 - 11997470 -----UCACAGUGUUACAACAUCAAAAAUCUUAAUAAGCGGCAUACCGCUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAAUGUGAG-- -----(((((.((((...((.(((.............(((((.....))))).....((.(((((.....))))).))....))).(((....)))...))..)))).))))).-- ( -26.30, z-score = -2.32, R) >anoGam1.chr3L 36990725 111 - 41284009 ---UCCUCUUGCAGUACAACGGCCAAAAUCGUCAUCAGUGGUAUACCGCAGCACGCCAAGUUCGACGAUAUCGAACCGUUGCUGAAGCCGUACGGUAAGGUGGAGCACUGCGAG-- ---....((((((((....(.(((...((((((....((((....)))).(.((.....)).))))))).....(((((.((....))...)))))..))).)...))))))))-- ( -36.20, z-score = -1.44, R) >apiMel3.Group12 8436621 111 - 9182753 -----UUACAGUACUUCAUCCUCAAAGGUAAUCGUGAACAACUUCCCAGCUCAGGCCAGAAUAGAAGAUAUCGAAGCUUUAUUUUCGAACUGUGGACAAGUGCAGUCCGUUGAAAA -----..(((((.((((..((.....)).............((..((......))..))....))))...((((((......)))))))))))((((.......))))........ ( -19.80, z-score = 0.66, R) >triCas2.ChLG5 4269814 114 - 18847211 UCGGUAUGUUGCAGUUCAACAUCAAAAAUUAUUAUAUCCAAUUUGGGUCCAAACGUCCGGUUCGAAGACAUCGAACAUCUACUUUUACAACACGGAGCUGUCGUUGCAUGUGAU-- ....(((((.(((((((..................((((.....))))...........((((((.....))))))..................)))))))....)))))....-- ( -21.20, z-score = 0.70, R) >consensus _____UCACAGUGUUACGACAUCAAAAAUCUUAAUAAGCGGCAUACCGAUGCAAACACGUUUCGAAGACAUCGAACCGCUACUGAAGCCCUAUGGCAUCGUCAAACAAUGUGAG__ .....((((...(((((((.................((((((........))........(((((.....))))).))))......(((....))).))))..)))...))))... (-11.19 = -11.81 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:13 2011