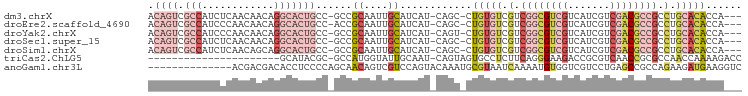
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,691,231 – 10,691,324 |
| Length | 93 |
| Max. P | 0.989158 |

| Location | 10,691,231 – 10,691,324 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 63.16 |
| Shannon entropy | 0.70561 |
| G+C content | 0.59495 |
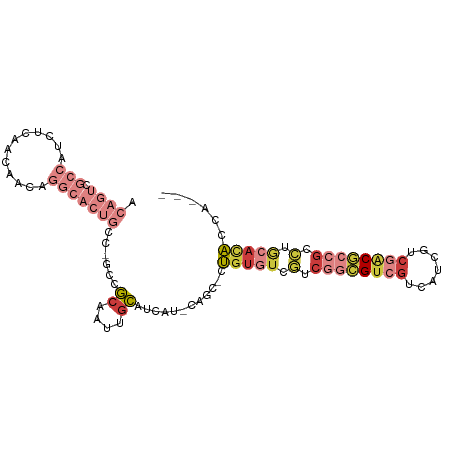
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -17.87 |
| Energy contribution | -18.51 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
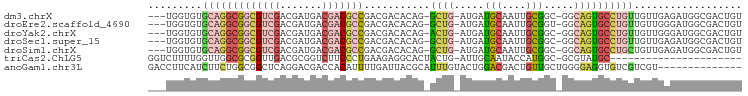
>dm3.chrX 10691231 93 + 22422827 ---UGGUGUGCAGGCGGCGUCGACGAUGACGACGCCGACGACACAG-GCUG-AUGAUGCAAUUGCGGC-GGCAGUGCCUGUUGUUGAGAUGGCGACUGU ---.(((.(((...((((((((.......)))))))).((((((((-((..-.((.(((.......))-).))..))))).))))).....)))))).. ( -37.10, z-score = -1.19, R) >droEre2.scaffold_4690 14309493 93 - 18748788 ---UGGUGUGCAGGCGGCGUCGACGAUGACGACGCCGACGACACAG-GCUG-AUGAUGCAAUUGCGGU-GGCAGUGCCUGUUGUUGGGAUGGCGACUGU ---.(((.(((...((((((((.......)))))))).((((((((-((..-.((.(((....)))..-..))..))))).))))).....)))))).. ( -37.30, z-score = -1.31, R) >droYak2.chrX 19261068 93 + 21770863 ---UGGUGUGCAGGCGGCGUCGACGAUGACGACGCCGACGACACAG-ACUG-AUGAUGCAAUUGCGGC-GGCAGUGCCUGUUGUUGGGAUGGCGACUGU ---...((((..(.((((((((.......)))))))).)..)))).-....-....(((.(((.((((-(((((...))))))))).))).)))..... ( -35.60, z-score = -0.93, R) >droSec1.super_15 1388688 93 + 1954846 ---UGGUGUGCAGGCGGCGUCGACGAUGACGACGCCGACGACACAG-GCUG-AUGAUGCAAUUGCGGC-GGCAGUGCCUGUUGUUGAGAUGGCGACUGU ---.(((.(((...((((((((.......)))))))).((((((((-((..-.((.(((.......))-).))..))))).))))).....)))))).. ( -37.10, z-score = -1.19, R) >droSim1.chrX 8311594 93 + 17042790 ---UGGUGUGCAGGCGGCGUCGACGAUGACGACGCCGACGACACAG-GCUG-AUGAUGCAAUUGCGGC-GGCAGUGCCUGCUGUUGAGAUGGCGACUGU ---.(((.(((...((((((((.......)))))))).((((((((-((..-.((.(((.......))-).))..))))).))))).....)))))).. ( -37.10, z-score = -1.06, R) >triCas2.ChLG5 4262422 75 + 18847211 GGUCUUUUGGUUGGCGCGGUUGACGCGGUCUUCCCUGAAGAGGCACUACUG-AUUGCAAUACCAUGGC-GCGUAUGC---------------------- .(((...((((..(((((.....)))(((((((......)))).)))....-...))...)))).)))-........---------------------- ( -19.30, z-score = 1.11, R) >anoGam1.chr3L 36986892 85 + 41284009 GACCUUCAUCUUCUGGCGGCUCAGGACGACCACAUUUUGAUUACGCAUUUGUACUGGACGACUGUUGCUGGGGAGGUGUCGUCGU-------------- (((...((((((((..((((.(((..((.(((((...((......))..)))...)).)).)))..))))))))))))..)))..-------------- ( -23.50, z-score = -0.01, R) >consensus ___UGGUGUGCAGGCGGCGUCGACGAUGACGACGCCGACGACACAG_GCUG_AUGAUGCAAUUGCGGC_GGCAGUGCCUGUUGUUGAGAUGGCGACUGU .........(((((((((((((.......)))))))...........((((.....(((....))).....)))))))))).................. (-17.87 = -18.51 + 0.64)
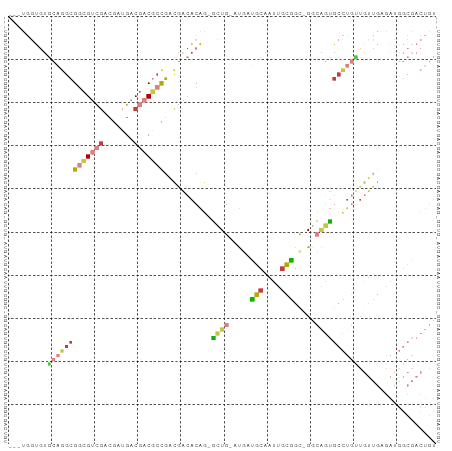
| Location | 10,691,231 – 10,691,324 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 63.16 |
| Shannon entropy | 0.70561 |
| G+C content | 0.59495 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -14.19 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
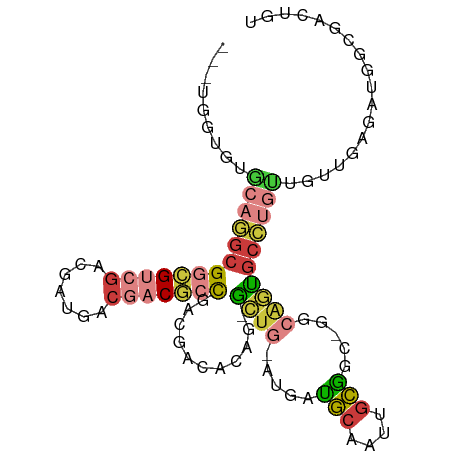
>dm3.chrX 10691231 93 - 22422827 ACAGUCGCCAUCUCAACAACAGGCACUGCC-GCCGCAAUUGCAUCAU-CAGC-CUGUGUCGUCGGCGUCGUCAUCGUCGACGCCGCCUGCACACCA--- .((((.(((............)))))))..-...((....)).....-....-.(((((.(.((((((((.......)))))))).).)))))...--- ( -30.30, z-score = -1.76, R) >droEre2.scaffold_4690 14309493 93 + 18748788 ACAGUCGCCAUCCCAACAACAGGCACUGCC-ACCGCAAUUGCAUCAU-CAGC-CUGUGUCGUCGGCGUCGUCAUCGUCGACGCCGCCUGCACACCA--- .((((.(((............)))))))..-...((....)).....-....-.(((((.(.((((((((.......)))))))).).)))))...--- ( -30.30, z-score = -2.38, R) >droYak2.chrX 19261068 93 - 21770863 ACAGUCGCCAUCCCAACAACAGGCACUGCC-GCCGCAAUUGCAUCAU-CAGU-CUGUGUCGUCGGCGUCGUCAUCGUCGACGCCGCCUGCACACCA--- .((((.(((............)))))))..-...((....)).....-....-.(((((.(.((((((((.......)))))))).).)))))...--- ( -30.30, z-score = -2.00, R) >droSec1.super_15 1388688 93 - 1954846 ACAGUCGCCAUCUCAACAACAGGCACUGCC-GCCGCAAUUGCAUCAU-CAGC-CUGUGUCGUCGGCGUCGUCAUCGUCGACGCCGCCUGCACACCA--- .((((.(((............)))))))..-...((....)).....-....-.(((((.(.((((((((.......)))))))).).)))))...--- ( -30.30, z-score = -1.76, R) >droSim1.chrX 8311594 93 - 17042790 ACAGUCGCCAUCUCAACAGCAGGCACUGCC-GCCGCAAUUGCAUCAU-CAGC-CUGUGUCGUCGGCGUCGUCAUCGUCGACGCCGCCUGCACACCA--- ..................((((((...(.(-(.((((...((.....-..))-.)))).)).)(((((((.......)))))))))))))......--- ( -32.20, z-score = -1.91, R) >triCas2.ChLG5 4262422 75 - 18847211 ----------------------GCAUACGC-GCCAUGGUAUUGCAAU-CAGUAGUGCCUCUUCAGGGAAGACCGCGUCAACCGCGCCAACCAAAAGACC ----------------------......((-((...((((((((...-..)))))))).......((..(((...)))..))))))............. ( -19.60, z-score = -0.93, R) >anoGam1.chr3L 36986892 85 - 41284009 --------------ACGACGACACCUCCCCAGCAACAGUCGUCCAGUACAAAUGCGUAAUCAAAAUGUGGUCGUCCUGAGCCGCCAGAAGAUGAAGGUC --------------..((((((...............))))))(((.((...(((((.......)))))...)).))).(((..((.....))..))). ( -14.56, z-score = 1.00, R) >consensus ACAGUCGCCAUCUCAACAACAGGCACUGCC_GCCGCAAUUGCAUCAU_CAGC_CUGUGUCGUCGGCGUCGUCAUCGUCGACGCCGCCUGCACACCA___ .....................(((.......)))((....))............(((((.(.((((((((.......)))))))).).)))))...... (-14.19 = -15.83 + 1.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:12 2011