| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,926,718 – 10,926,810 |
| Length | 92 |
| Max. P | 0.935670 |

| Location | 10,926,718 – 10,926,810 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Shannon entropy | 0.32847 |
| G+C content | 0.45406 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -19.10 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
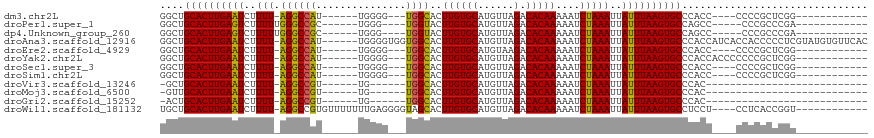
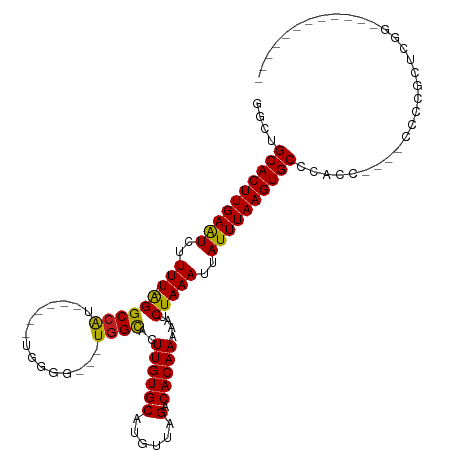
>dm3.chr2L 10926718 92 + 23011544 GGCUGCACUUGAAUCUUUU-AGGCCAU------UGGGG---UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACC----CCCCGCUCGG------------ ((((...............-.))))..------.((((---(((((((((......(((((........)))))......)))))).)))))----))........------------ ( -29.39, z-score = -2.07, R) >droPer1.super_1 758755 91 + 10282868 GGCUGCACUUGAGUCUUUUUGGGCCGC------UGGG----UGGUACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCAGCC-----CCCGCCCGA------------ ((((.......))))...((((((...------.(((----(((((((((......(((((........)))))......))))))))).))-----)..))))))------------ ( -30.50, z-score = -1.79, R) >dp4.Unknown_group_260 43546 91 + 94298 GGCUGCACUUGAGUCUUUUUGGGCCGC------UGGG----UGGUACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCAGCC-----CCCGCCCGA------------ ((((.......))))...((((((...------.(((----(((((((((......(((((........)))))......))))))))).))-----)..))))))------------ ( -30.50, z-score = -1.79, R) >droAna3.scaffold_12916 14784935 111 + 16180835 GGCUGCACUUGAAUCUUUU-AGGCCAU------UGGGGUGGUGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACCAUCACCACCCCCUCGUAUGUGUUCAC ((((...............-.))))..------.((((((((((((((((......(((((........)))))......)))))))).......))))))))............... ( -33.60, z-score = -1.69, R) >droEre2.scaffold_4929 12128956 92 - 26641161 GGCUGCACUUGAAUCUUUU-AGGCCAU------UGGGG---UGGCACUUGUGCAUGUAAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACC----CCCCGCUCGG------------ ((..((((((((((..(((-(((((((------....)---))))..((((((......).)))))....)))))..))))))))))))...----..........------------ ( -28.10, z-score = -1.66, R) >droYak2.chr2L 7326729 96 + 22324452 GGCUGCACUUGAAUCUUUU-AGGCCAU------UGGGG---UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACCACCCCCCCGCUCGG------------ ((..((((((((((..(((-(((((((------....)---))))..((((((......).)))))....)))))..))))))))))...))..............------------ ( -28.60, z-score = -1.50, R) >droSec1.super_3 6337746 92 + 7220098 GGCUGCACUUGAAUCUUUU-AGGCCAU------UGGGG---UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACC----CCCCGCUCGG------------ ((((...............-.))))..------.((((---(((((((((......(((((........)))))......)))))).)))))----))........------------ ( -29.39, z-score = -2.07, R) >droSim1.chr2L 10727130 92 + 22036055 GGCUGCACUUGAAUCUUUU-AGGCCAU------UGGGG---UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACC----CCCCGCUCGG------------ ((((...............-.))))..------.((((---(((((((((......(((((........)))))......)))))).)))))----))........------------ ( -29.39, z-score = -2.07, R) >droVir3.scaffold_13246 1272368 77 + 2672878 -GCUGCACUUGAAUCUUUU-AGGCCGU------UG------UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCAC--------------------------- -...((((((((((..(((-(((((..------..------.)))..((((((......).)))))....)))))..))))))))))....--------------------------- ( -23.70, z-score = -3.21, R) >droMoj3.scaffold_6500 27723422 77 - 32352404 -GUUGCACUUGAAUCUUUU-AGGCCGU------UG------UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCAC--------------------------- -...((((((((((..(((-(((((..------..------.)))..((((((......).)))))....)))))..))))))))))....--------------------------- ( -23.70, z-score = -3.32, R) >droGri2.scaffold_15252 1246423 77 - 17193109 -ACUGCACUUGAAUCUUUU-AGGCCGU------UG------UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCAC--------------------------- -...((((((((((..(((-(((((..------..------.)))..((((((......).)))))....)))))..))))))))))....--------------------------- ( -23.70, z-score = -3.52, R) >droWil1.scaffold_181132 546151 101 + 1035393 UGCUGCACUUGAAUCUUUU-AGGCCGUGUUUUUUUGAGGGGUAGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCUCCU----CCUCACCGGU------------ ...................-..((((.((......((((((..(((((((......(((((........)))))......)))))))..)).----))))))))))------------ ( -26.30, z-score = -1.32, R) >consensus GGCUGCACUUGAAUCUUUU_AGGCCAU______UGGGG___UGGCACUUGUGCAUGUUAGACACAAAAAUCUAAAUUAUUUAAGUGCCCACC____CCCCGCUCGG____________ ....((((((((((..(((.((((((...............))))..((((((......).)))))....)))))..))))))))))............................... (-19.10 = -18.52 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:13 2011