
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,644,642 – 10,644,784 |
| Length | 142 |
| Max. P | 0.766153 |

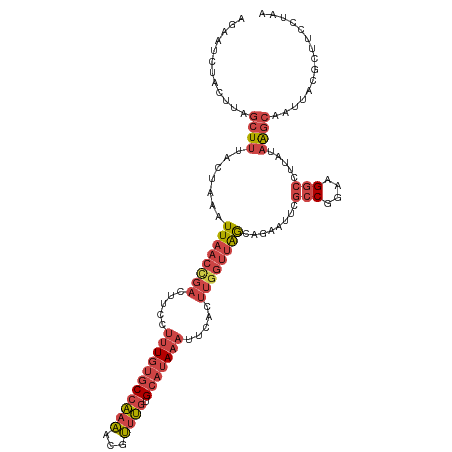
| Location | 10,644,642 – 10,644,759 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.46215 |
| G+C content | 0.38204 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -14.69 |
| Energy contribution | -16.53 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.766153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10644642 117 + 22422827 AGAAUCUACUUAGCUUUACUAAAUUAACCGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAGCAGAAUUUGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAA ............((((.......((((((((.....((((((((((.....))).))))))).....))))))))........(((....))).....))))............... ( -24.90, z-score = -1.35, R) >droSim1.chrX 8273937 114 + 17042790 AGAAUCUACUUAGCUUUACUAAAUUAACCGACUUCCUUUGUGCCAAA-CGUUUGUGCAUAAAUUCACUUGGUUAGCAGA-CUCGCCGGAAGGC-UUAUAAGCAAUUACGCUUCCUAA .((.(((..((((.....)))).((((((((.....((((((((((.-...))).))))))).....)))))))).)))-.))(((....)))-....((((......))))..... ( -27.60, z-score = -2.25, R) >droSec1.super_15 1343236 117 + 1954846 AGAAUCUACUUAGCUUUACUAAAUUAACCGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAACAGAAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAA .((((((..((((.....)))).((((((((.....((((((((((.....))).))))))).....)))))))).)).))))(((....))).....((((......))))..... ( -27.40, z-score = -2.81, R) >droYak2.chrX 19213495 117 + 21770863 AGAAUCUACUUAGCUUCACGAAAUUAACUGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAACAGCAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCCAA ............((((...(...(((((..(.....((((((((((.....))).))))))).....)..)))))...)....(((....))).....))))............... ( -23.10, z-score = -0.80, R) >droEre2.scaffold_4690 14262983 117 - 18748788 AGAAUCUACUUAGCUUUACGAAAUUAACCGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAGCAGAAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAA ............((((.......((((((((.....((((((((((.....))).))))))).....))))))))........(((....))).....))))............... ( -25.20, z-score = -1.21, R) >droVir3.scaffold_12932 546465 97 - 2102469 AAAGAUUGCCUAAAUUCGUCAAUUU---CUACAUCGUCUAUGCUGCUUAGAUC-UGAUCAACAUGAUGUUAUCGGUUUACUAUAACAUAUGGCUUUACUGG---------------- .......(((...............---...((..(((((.......))))).-)).........(((((((.(.....).)))))))..)))........---------------- ( -12.20, z-score = 0.89, R) >consensus AGAAUCUACUUAGCUUUACUAAAUUAACCGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAGCAGAAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAA ............((((.......((((((((.....(((((((((((...)))).))))))).....))))))))........(((....))).....))))............... (-14.69 = -16.53 + 1.84)


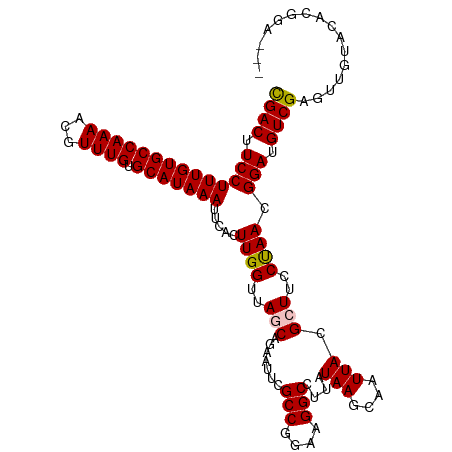
| Location | 10,644,670 – 10,644,784 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.69 |
| Shannon entropy | 0.09106 |
| G+C content | 0.44557 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -28.10 |
| Energy contribution | -28.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10644670 114 + 22422827 CGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAGCAGAAUUUGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAACGGAUGUCGAGUUGUACACGGAAAA ((((.(((((((((((((.....))).)))))))........(((((........(((....))).....((((......)))).)))))))).))))................ ( -30.40, z-score = -1.37, R) >droSim1.chrX 8273965 107 + 17042790 CGACUUCCUUUGUGCCAAA-CGUUUGUGCAUAAAUUCACUUGGUUAGCAGA-CUCGCCGGAAGGC-UUAUAAGCAAUUACGCUUCCUAACGGAUGUCGAGUUGUACACGG---- ((((.(((((((((((((.-...))).)))))))........(((((....-...(((....)))-....((((......)))).)))))))).))))............---- ( -31.10, z-score = -1.87, R) >droSec1.super_15 1343264 114 + 1954846 CGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAACAGAAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAACGGAUGUCGAGUUGUACACGGAAAA ....((((((((((((((.....))).))))))).....................(((....))).......((((((.(((.(((....))).).))))))))....)))).. ( -29.00, z-score = -1.24, R) >droYak2.chrX 19213523 111 + 21770863 UGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAACAGCAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCCAACGGAUGUCGAGUUGUACACGGA--- .(((.(((((((((((((.....))).))))))).....((((............(((....))).....((((......)))).)))).))).)))..............--- ( -27.30, z-score = -0.43, R) >droEre2.scaffold_4690 14263011 111 - 18748788 CGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAGCAGAAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAACGGAUGUCGAGUUGUACACGGA--- ((((.(((((((((((((.....))).)))))))........(((((........(((....))).....((((......)))).)))))))).)))).............--- ( -30.70, z-score = -1.42, R) >consensus CGACUUCCUUUGUGCCAAAACGUUUGUGCAUAAAUUCACUUGGUUAGCAGAAUUCGCCGGAAGGCCUUAUAAGCAAUUACGCUUCCUAACGGAUGUCGAGUUGUACACGGA___ ((((.(((((((((((((.....))).)))))))........(((((........(((....))).....((((......)))).)))))))).))))................ (-28.10 = -28.54 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:07 2011