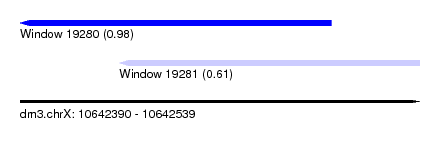
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,642,390 – 10,642,539 |
| Length | 149 |
| Max. P | 0.982770 |

| Location | 10,642,390 – 10,642,506 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.20 |
| Shannon entropy | 0.22793 |
| G+C content | 0.41977 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -24.78 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
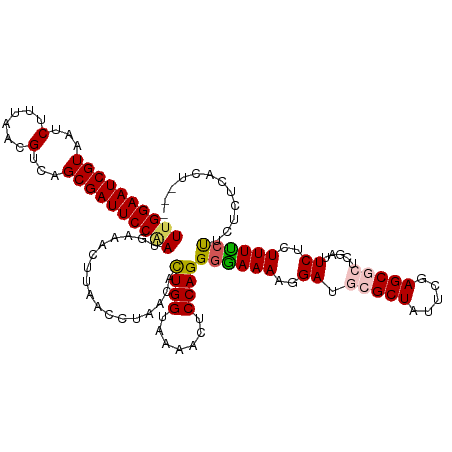
>dm3.chrX 10642390 116 - 22422827 UUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCGAUUAAACUUAACCUAACAUUGGAAAAAAUCCAGGGGAAAAGGAUGCGCUAUUCGAGCGCUCGAUUCUCUUUUCUUCUCUCACU--- ((((((((((...(......)...))))))))))..................((((.....))))(((((((((((.(((((.....))))).....)).))))))))).......--- ( -32.40, z-score = -2.93, R) >droSim1.chrX 8271723 116 - 17042790 UUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCAAUGAAACUUAACCUAACACUGGUAAAAUUCCAGGGAAAAAGGAUGCGCUAUUCGAGCGCUCGAUUCACUUUUCUUCUCUCACU--- ((((((((((...(......)...))))))))))(((..............((((.......))))((((((..((.(((((.....))))).....))..))))))....)))..--- ( -29.30, z-score = -2.72, R) >droSec1.super_15 1341031 116 - 1954846 UUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCAAUGAAACUUAACCUAACACUGGAAAAAUUCCAGGGAAAAAGGAUGCGCUAUUCGAGCGCCCGAUUCACUUUUCUUCUCUCACU--- ((((((((((...(......)...))))))))))(((..............(((((.....)))))((((((..((.(((((.....))))).....))..))))))....)))..--- ( -30.10, z-score = -2.78, R) >droYak2.chrX 19211206 119 - 21770863 UUGGAAUCGUUAUCUUUAACGUCGGCGAUUCCAACGAGACUUAACCUAACACUGGUAAAACUCCAGGGGAAAAGGAUGCGCUAUUCGAGCAAUCGAUUCUCUUUUCUUCUCUUCUUACU (((((((((((..(......)..))))))))))).((((............((((.......)))).(((((((((........((((....)))).)).)))))))))))........ ( -32.30, z-score = -2.68, R) >droEre2.scaffold_4690 14260807 110 + 18748788 UUGGAAUCGUAAGCUUUAACGUCGGCGAUUCCUACGAGACUUAACCUAACACUGGUAAAACUCCAGGGGAAAAGGACGCGCUAUUCGAGC----GAUUCUCUUUCACUUUCACU----- ..((((((((..((......))..))))))))...((((............((((.......))))..((((..((..((((.....)))----)..))..))))..))))...----- ( -30.70, z-score = -2.17, R) >consensus UUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCAAUGAAACUUAACCUAACACUGGUAAAACUCCAGGGGAAAAGGAUGCGCUAUUCGAGCGCUCGAUUCUCUUUUCUUCUCUCACU___ ((((((((((...(......)...)))))))))).................((((.......))))((((((..((.(((((.....))))).....))..))))))............ (-24.78 = -25.14 + 0.36)

| Location | 10,642,427 – 10,642,539 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.27 |
| Shannon entropy | 0.18722 |
| G+C content | 0.42522 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.76 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
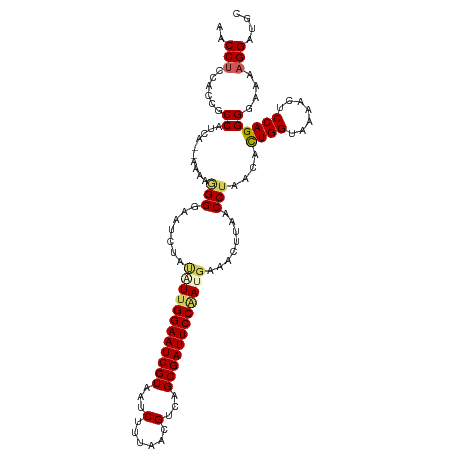
>dm3.chrX 10642427 112 - 22422827 AACCUCCACCGCCAUCA--AAAAGGGGAAUCUACAUUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCGAUUAAACUUAACCUAACAUUGGAAAAAAUCCAGGGGAAAAGGAUGC ..(((...((.((....--...(((..((.....(((((((((((...(......)...))))))))))).....))..))).....((((.....))))))))...))).... ( -27.30, z-score = -1.66, R) >droSim1.chrX 8271760 112 - 17042790 AACCUCCACCGCCAUCA--AAAAGGGGAAUCUACGUUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCAAUGAAACUUAACCUAACACUGGUAAAAUUCCAGGGAAAAAGGAUGC ....(((.((.((....--....))))..(((.((((((((((((...(......)...))))))))))))...............((((.......)))))))....)))... ( -29.20, z-score = -2.33, R) >droSec1.super_15 1341068 112 - 1954846 AACCCUCACCGCCAUCA--AAAAGGGGAAUAUAUGUUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCAAUGAAACUUAACCUAACACUGGAAAAAUUCCAGGGAAAAAGGAUGC ...(((.....((....--...(((..((.....(((((((((((...(......)...))))))))))).....))..)))....(((((.....)))))))....))).... ( -28.30, z-score = -2.19, R) >droYak2.chrX 19211246 114 - 21770863 AACCUCCACCGCCACCAAAAAAAAGGGAAUCUAUAUUGGAAUCGUUAUCUUUAACGUCGGCGAUUCCAACGAGACUUAACCUAACACUGGUAAAACUCCAGGGGAAAAGGAUGC ..(((...((.............(((...(((...(((((((((((..(......)..)))))))))))..))).....)))....((((.......))))))....))).... ( -28.30, z-score = -1.46, R) >droEre2.scaffold_4690 14260838 112 + 18748788 AACCUGCAAGGCCACCA--AAAAUGGGAAUCUAUAUUGGAAUCGUAAGCUUUAACGUCGGCGAUUCCUACGAGACUUAACCUAACACUGGUAAAACUCCAGGGGAAAAGGACGC ..(((......((....--....((((..(((.....((((((((..((......))..))))))))....))).....))))...((((.......)))).))...))).... ( -27.40, z-score = -0.95, R) >consensus AACCUCCACCGCCAUCA__AAAAGGGGAAUCUAUAUUGGAAUCGUAAUCUUUAACGUCAGCGAUUCCAAUGAAACUUAACCUAACACUGGUAAAACUCCAGGGGAAAAGGAUGC ..(((......((..........(((.......((((((((((((...(......)...))))))))))))........)))....((((.......))))))....))).... (-22.16 = -22.76 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:06 2011