| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,641,230 – 10,641,297 |
| Length | 67 |
| Max. P | 0.981354 |

| Location | 10,641,230 – 10,641,297 |
|---|---|
| Length | 67 |
| Sequences | 5 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 70.01 |
| Shannon entropy | 0.52107 |
| G+C content | 0.28505 |
| Mean single sequence MFE | -11.75 |
| Consensus MFE | -5.76 |
| Energy contribution | -5.40 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10641230 67 - 22422827 UUCAUCGCGACGUAAUUAGCCAUAGAUAAUGGUUAAAAAUAGUUUUGAAAACGUGAGUAUAUCAAAA-- ....(((.(((....((((((((.....)))))))).....))).)))..((....)).........-- ( -12.70, z-score = -1.99, R) >droSim1.chrX 8270593 55 - 17042790 UUCAUCGCGACAUAAUUAACUAUAGAUAAUGGUUGAUAAUUG-UCUAAAAACUAAA------------- ........((((..(((((((((.....)))))))))...))-))...........------------- ( -10.80, z-score = -3.31, R) >droSec1.super_15 1339913 55 - 1954846 UUCAUCGCGACAUAAUUAACUAUAGAUAAUGGUUGAUAAUUG-UCUAAAAACUAAA------------- ........((((..(((((((((.....)))))))))...))-))...........------------- ( -10.80, z-score = -3.31, R) >droYak2.chrX 19210062 68 - 21770863 CUAAUCGCGACGUAAUUAGCCAUAGAUAAUGGUUAAUAAUUG-UCUGAAAAGUAAGUUAGAGGACUUAA ....(((.((((..(((((((((.....)))))))))...))-)))))....((((((....)))))). ( -16.80, z-score = -3.18, R) >droEre2.scaffold_4690 14259699 68 + 18748788 UUCGUCGCGACGUAAUUGGCCAUAGAUAGUCAUUAAUAAAUG-UAUAAAAAGAAAAUUAUAUGACUGUA ..(((....))).............((((((((((((.....-............)))).)))))))). ( -7.63, z-score = 0.50, R) >consensus UUCAUCGCGACGUAAUUAGCCAUAGAUAAUGGUUAAUAAUUG_UCUAAAAACUAAA_UA_A__A_____ ..............(((((((((.....)))))))))................................ ( -5.76 = -5.40 + -0.36)

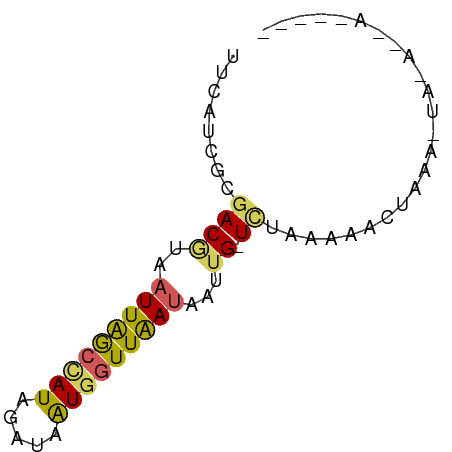

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:04 2011