| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,593,232 – 10,593,326 |
| Length | 94 |
| Max. P | 0.556964 |

| Location | 10,593,232 – 10,593,326 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.59745 |
| G+C content | 0.38174 |
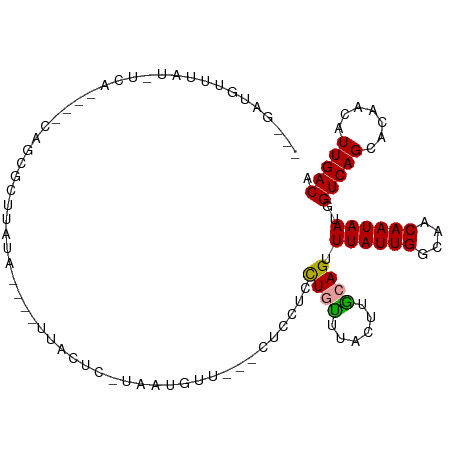
| Mean single sequence MFE | -17.12 |
| Consensus MFE | -11.10 |
| Energy contribution | -10.93 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
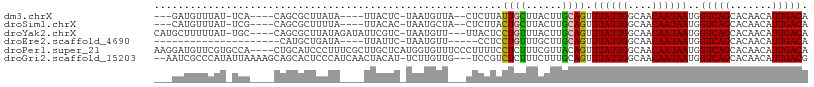
>dm3.chrX 10593232 94 + 22422827 ---GAUGUUUAU-UCA----CAGCGCUUAUA----UUACUC-UAAUGUUA--CUCUUAUUGCUUACUUGCAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACA ---.........-...----...........----......-(((((((.--.....(((((......)))))((((((....))))))..........)))))))... ( -14.30, z-score = 0.24, R) >droSim1.chrX 8220575 94 + 17042790 ---CAUGUUUAU-UCG----CAGCGCUUUUA----UUACAC-UAAUGCUA--CUCUUACUGCUUACUUGCAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACA ---.(((((...-..(----(...)).....----......-...(((((--((...(((((......)))))((((((....)))))).))).)))).)))))..... ( -17.70, z-score = -0.69, R) >droYak2.chrX 19159462 100 + 21770863 CAUGCUUUUUAU-UGC----CAGCGCUUAUAGAUAUUCGUC-UAAUGUU---UUACUCCUGUUUACUUGCAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACA .......(((((-.((----....))..))))).....(((-.((((((---......((((......)))).((((((....))))))..........))))))))). ( -17.60, z-score = 0.09, R) >droEre2.scaffold_4690 14211399 78 - 18748788 ---------------------CAUGCUGAUA----UUAUUC-UAAUGUU-----CCUCCUGUUUGCUUGCAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACA ---------------------..(((((((.----......-.......-----....((((......)))).((((((....))))))..)))))))........... ( -18.70, z-score = -1.51, R) >droPer1.super_21 918687 105 + 1645244 AAGGAUGUUCGUGCCA----CUGCAUCCCUUUCGCUUGCUCAUGGUGUUUCCCUUUUCCUCUUUCGUUACAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACA ...((((((.((((((----(((((...........)))....)))).....................((...((((((....))))))..))..)))))))))).... ( -20.20, z-score = 0.08, R) >droGri2.scaffold_15203 7168207 103 + 11997470 --AAUCGCCCAUAUUAAAAGCAGCACUCCCAUCAACUACAU-UCUUGUUG---UCCGUCUCUUUCUUUGCAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACG --....................................(((-(.((((((---((.((..((........))...)).)))))))).))))(((((.......))))). ( -14.20, z-score = 0.09, R) >consensus ___GAUGUUUAU_UCA____CAGCGCUUAUA____UUACUC_UAAUGUU___CUCCUCCUGUUUACUUGCAGUUUAUUGGCAACAAUAAUGGUCAGCACAACAUUGACA ..........................................................((((......)))).((((((....))))))..(((((.......))))). (-11.10 = -10.93 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:58 2011