
| Sequence ID | dm3.chrX |
|---|---|
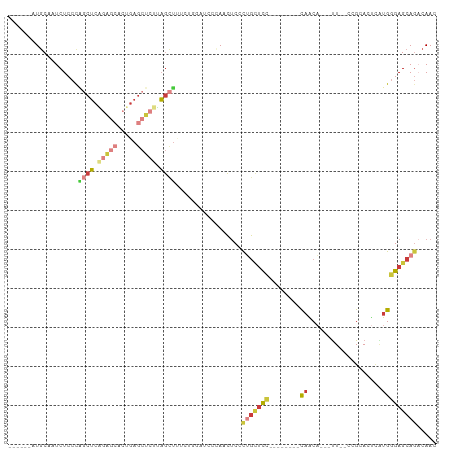
| Location | 10,568,282 – 10,568,373 |
| Length | 91 |
| Max. P | 0.983234 |

| Location | 10,568,282 – 10,568,373 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 63.34 |
| Shannon entropy | 0.67069 |
| G+C content | 0.53714 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -9.14 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
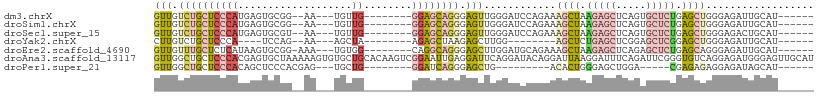
>dm3.chrX 10568282 91 + 22422827 ------AUGCAAUCUCCCAGCUCAGAGCACUGAGCUCUUAGCUUUCUGGAUCCCAACUCCCUGCUCC--------CAACA---UU--CCGCACUCAUGGGAGCAGACAAC ------.((..((((...((((.(((((.....))))).))))....))))..)).....(((((((--------((...---..--.........)))))))))..... ( -30.24, z-score = -3.56, R) >droSim1.chrX 8200287 91 + 17042790 ------AUGCAAUCUCCCAGCUCAGAGCACUGAGCUCUUAGCUUUCUGGAUCCCAACUCCCUGCUCC--------CAACA---UU--CCGCACUCAUGGGAGCAGACAAC ------.((..((((...((((.(((((.....))))).))))....))))..)).....(((((((--------((...---..--.........)))))))))..... ( -30.24, z-score = -3.56, R) >droSec1.super_15 1272647 91 + 1954846 ------AUGCAGUCUCCCAGCUCAGAGCACUGAGCUCUUAGCUUUCUGGAUCCCAACUCCCUGCUCC--------CAACA---UU--ACGCACUCAUGGGAGCAGACAAC ------.((..((((...((((.(((((.....))))).))))....))))..)).....(((((((--------((...---..--.........)))))))))..... ( -30.54, z-score = -3.18, R) >droYak2.chrX 19137753 79 + 21770863 ------AUGCAAUCUCCCAGCUCCGAGCUCCGAGCUCAGAGCU--------CCAAGCUCUUAGCUCU--------UAGCU---UU--CUGGA----UGGGAGCAGACAAG ------.......((((((..((((((((..(((((.(((((.--------....))))).))))).--------.))))---).--..)))----))))))........ ( -33.40, z-score = -2.69, R) >droEre2.scaffold_4690 14192537 92 - 18748788 ------AUGCAAUCUCCCUGCUCAGAGCUCUGAGCUCUUAGCUUUCUGCAUCCAAGCUCCCUGCCUG--------CCACA---UUU-CCGCACUUAUGAGAGCAAACAAC ------(((((........(((.(((((.....))))).)))....)))))....(((((.((..((--------(....---...-..)))..)).).))))....... ( -20.50, z-score = -0.82, R) >droAna3.scaffold_13117 4585376 110 + 5790199 AUGCAACUCCCAUCUCCUGACACCCGAAUCUGAAAUCCUUAAUCCUGUAUCCUGAAUCCUCAAUUCCGACUUGUGCAGCACACUUUUUAGCACUCGUGGGAGCAGCCAAC ..((..((((((((....))....(((..((((((.........((((((.(.((((.....)))).)....))))))......))))))...)))))))))..)).... ( -19.26, z-score = -0.60, R) >droPer1.super_21 896729 79 + 1645244 ------AUGCUAUCUCCUCUCUCG-----UCCAGCUCCCAGUGU---------CAGCUCCCUGAUCC--------CAGCA---CUCGUGGGAGCUGUGGGAGCAGCCAAC ------..................-----....(((((((....---------(((((((((((...--------.....---.))).)))))))))))))))....... ( -29.10, z-score = -1.93, R) >consensus ______AUGCAAUCUCCCAGCUCAGAGCACUGAGCUCUUAGCUUUCUGGAUCCCAACUCCCUGCUCC________CAACA___UU__CCGCACUCAUGGGAGCAGACAAC .............(((((((((.(((((.....))))).)))).............(.....)..................................)))))........ ( -9.14 = -9.90 + 0.76)


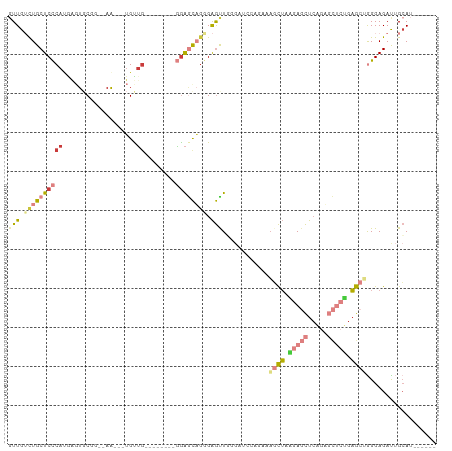
| Location | 10,568,282 – 10,568,373 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 63.34 |
| Shannon entropy | 0.67069 |
| G+C content | 0.53714 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -14.19 |
| Energy contribution | -15.26 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
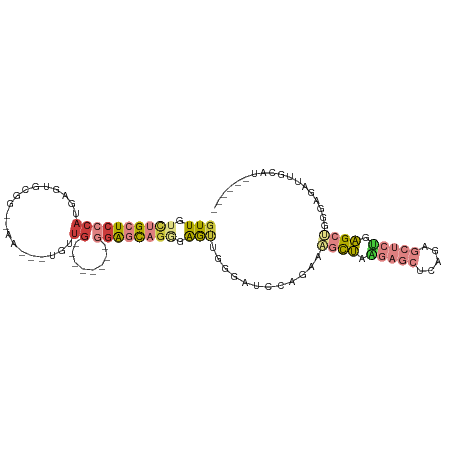
>dm3.chrX 10568282 91 - 22422827 GUUGUCUGCUCCCAUGAGUGCGG--AA---UGUUG--------GGAGCAGGGAGUUGGGAUCCAGAAAGCUAAGAGCUCAGUGCUCUGAGCUGGGAGAUUGCAU------ .((.((((((((((...((....--.)---)..))--------)))))))).)).((.(((((....((((.(((((.....))))).))))..).)))).)).------ ( -37.40, z-score = -2.64, R) >droSim1.chrX 8200287 91 - 17042790 GUUGUCUGCUCCCAUGAGUGCGG--AA---UGUUG--------GGAGCAGGGAGUUGGGAUCCAGAAAGCUAAGAGCUCAGUGCUCUGAGCUGGGAGAUUGCAU------ .((.((((((((((...((....--.)---)..))--------)))))))).)).((.(((((....((((.(((((.....))))).))))..).)))).)).------ ( -37.40, z-score = -2.64, R) >droSec1.super_15 1272647 91 - 1954846 GUUGUCUGCUCCCAUGAGUGCGU--AA---UGUUG--------GGAGCAGGGAGUUGGGAUCCAGAAAGCUAAGAGCUCAGUGCUCUGAGCUGGGAGACUGCAU------ .((.((((((((((...((....--.)---)..))--------)))))))).))..((..(((....((((.(((((.....))))).)))).)))..))....------ ( -38.70, z-score = -2.91, R) >droYak2.chrX 19137753 79 - 21770863 CUUGUCUGCUCCCA----UCCAG--AA---AGCUA--------AGAGCUAAGAGCUUGG--------AGCUCUGAGCUCGGAGCUCGGAGCUGGGAGAUUGCAU------ ..(((...((((((----.....--..---.(((.--------.(((((..(((((..(--------....)..)))))..)))))..)))))))))...))).------ ( -36.80, z-score = -2.53, R) >droEre2.scaffold_4690 14192537 92 + 18748788 GUUGUUUGCUCUCAUAAGUGCGG-AAA---UGUGG--------CAGGCAGGGAGCUUGGAUGCAGAAAGCUAAGAGCUCAGAGCUCUGAGCAGGGAGAUUGCAU------ ..(((...(((((......((..-...---((((.--------(((((.....)))))..))))....)).....((((((....)))))).)))))...))).------ ( -29.90, z-score = -0.51, R) >droAna3.scaffold_13117 4585376 110 - 5790199 GUUGGCUGCUCCCACGAGUGCUAAAAAGUGUGCUGCACAAGUCGGAAUUGAGGAUUCAGGAUACAGGAUUAAGGAUUUCAGAUUCGGGUGUCAGGAGAUGGGAGUUGCAU ....((.((((((((..(..((....))..).((((((.((((((((((...(((((........)))))...)))))).))))...))).)))..).))))))).)).. ( -31.60, z-score = -0.90, R) >droPer1.super_21 896729 79 - 1645244 GUUGGCUGCUCCCACAGCUCCCACGAG---UGCUG--------GGAUCAGGGAGCUG---------ACACUGGGAGCUGGA-----CGAGAGAGGAGAUAGCAU------ ....((((((((..(((((((((...(---((...--------...((((....)))---------)))))))))))))..-----(....).)))).))))..------ ( -33.50, z-score = -2.15, R) >consensus GUUGUCUGCUCCCAUGAGUGCGG__AA___UGUUG________GGAGCAGGGAGUUGGGAUCCAGAAAGCUAAGAGCUCAGAGCUCUGAGCUGGGAGAUUGCAU______ ..(((...(((((................................(((.....)))...........((((.(((((.....))))).)))))))))...)))....... (-14.19 = -15.26 + 1.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:56 2011