
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,535,464 – 10,535,599 |
| Length | 135 |
| Max. P | 0.933528 |

| Location | 10,535,464 – 10,535,566 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 58.66 |
| Shannon entropy | 0.77217 |
| G+C content | 0.36159 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -8.46 |
| Energy contribution | -8.23 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.95 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613910 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
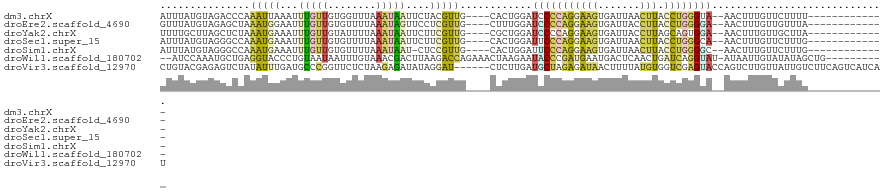
>dm3.chrX 10535464 102 + 22422827 AUUUAUGUAGACCCAAAUUAAAUUUGUUGUGGUUUAAAUAAUUCUACGUUG----CACUGGAUCCCCAGGAAGUGAUUAACUUACCUGGGUA--AACUUUGUUCUUUU------------- ....(((((((......(((((((......))))))).....))))))).(----((..(..(.((((((((((.....)))).)))))).)--..)..)))......------------- ( -23.40, z-score = -1.64, R) >droEre2.scaffold_4690 14158309 102 - 18748788 GUUUAUGUAGAGCUAAAUGGAAUUUGUUGUGUUUUAAAUAGUUCCUCGUUG----CUUUGGAUCCCCAGGAAGUGAUUACCUUACCUGGGGA--AACUUUGUUGUUUA------------- .....(.((((((.....((((((.(((........))))))))).....)----))))).)(((((((((((.......))).))))))))--..............------------- ( -29.10, z-score = -3.02, R) >droYak2.chrX 19104916 102 + 21770863 UUUUGCUUAGCUCUAAAUGAAAUUUGUUGUAUUUUAAAUAAUUCUUCGUUG----CGCUGGAUCCCCAGGAAGUGAUUACCUUAGCAGUGGA--AACUUUGUUGCUUA------------- ..(..(((.((....((((((..(((((........)))))...)))))))----).((((....)))).)))..)......((((((.(..--..).))))))....------------- ( -19.00, z-score = -0.13, R) >droSec1.super_15 1239016 102 + 1954846 AUUUAUGUAGGGCCAAAUGAAAUUUGUUGUGUUUUAAAUAAUUCUUCGUUG----CACUGGAUUCCCAGGAAGUGAUUAACUUACCUGGGCA--AACUUUGUUCUUUG------------- ....((.(((.((..((((((..(((((........)))))...)))))))----).))).)).((((((((((.....)))).))))))..--..............------------- ( -22.30, z-score = -1.00, R) >droSim1.chrX 8163757 101 + 17042790 AUUUAUGUAGGGCCAAAUGAAAUUUGUUGUGUUUUAAAUAAU-CUCCGUUG----CACUGGAUUCCCAGGAAGUGAUUAACUUACCUGGGGC--AACUUUGUUCUUUG------------- .............((((.(((....(((((............-.((((...----...))))..((((((((((.....)))).))))))))--)))....)))))))------------- ( -25.60, z-score = -1.57, R) >droWil1.scaffold_180702 3791076 108 + 4511350 --AUCCAAAUGCUGAGGUACCCUGUAAUAAUUUGUAAACGACUUAAGACCAGAAACUAAGAAUACCCGAUGAAUGACUCAACUGAUCAGGUAU-AUAAUUGUAUAUAGCUG---------- --........((((..((((.((((..(((.(((....))).)))..).))).........(((((.(((.(.(......).).))).)))))-......)))).))))..---------- ( -13.70, z-score = 0.41, R) >droVir3.scaffold_12970 4425622 115 - 11907090 CUGUACGAGAGUCUAUAUUUGAUGCCCGGUUCUCUAAGAGAUAUAGGAU------CUCUUGAUGCUAGAGAUAACUUUUAUGUGGUCGAGUACCAGUCUUGUUAUUGUCUUCAGUCAUCAU ......((((..((((((((..................))))))))..)------))).(((((((..((((((........((((.....)))).........))))))..)).))))). ( -25.40, z-score = 0.51, R) >consensus AUUUAUGUAGGGCCAAAUGAAAUUUGUUGUGUUUUAAAUAAUUCUUCGUUG____CACUGGAUCCCCAGGAAGUGAUUAACUUACCUGGGGA__AACUUUGUUCUUUG_____________ ...............(((((...(((((........)))))....)))))............(((((((((((.......))).))))))))............................. ( -8.46 = -8.23 + -0.23)


| Location | 10,535,464 – 10,535,566 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 58.66 |
| Shannon entropy | 0.77217 |
| G+C content | 0.36159 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -7.39 |
| Energy contribution | -7.03 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.78 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685757 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
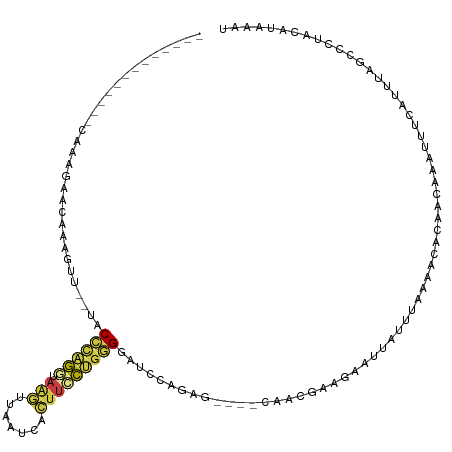
>dm3.chrX 10535464 102 - 22422827 -------------AAAAGAACAAAGUU--UACCCAGGUAAGUUAAUCACUUCCUGGGGAUCCAGUG----CAACGUAGAAUUAUUUAAACCACAACAAAUUUAAUUUGGGUCUACAUAAAU -------------..............--..((((((.((((.....))))))))))(((((((..----...................................)))))))......... ( -19.89, z-score = -1.23, R) >droEre2.scaffold_4690 14158309 102 + 18748788 -------------UAAACAACAAAGUU--UCCCCAGGUAAGGUAAUCACUUCCUGGGGAUCCAAAG----CAACGAGGAACUAUUUAAAACACAACAAAUUCCAUUUAGCUCUACAUAAAC -------------...........(..--((((((((.(((.......)))))))))))..)..((----(.....((((...................)))).....))).......... ( -22.41, z-score = -3.17, R) >droYak2.chrX 19104916 102 - 21770863 -------------UAAGCAACAAAGUU--UCCACUGCUAAGGUAAUCACUUCCUGGGGAUCCAGCG----CAACGAAGAAUUAUUUAAAAUACAACAAAUUUCAUUUAGAGCUAAGCAAAA -------------..(((.....(((.--......))).(((((((..((((((((....))))..----....)))).)))))))........................)))........ ( -10.80, z-score = 1.43, R) >droSec1.super_15 1239016 102 - 1954846 -------------CAAAGAACAAAGUU--UGCCCAGGUAAGUUAAUCACUUCCUGGGAAUCCAGUG----CAACGAAGAAUUAUUUAAAACACAACAAAUUUCAUUUGGCCCUACAUAAAU -------------((((.....(((((--((((((((.((((.....))))))))))......(((----....(((......)))....)))..)))))))..))))............. ( -19.10, z-score = -1.40, R) >droSim1.chrX 8163757 101 - 17042790 -------------CAAAGAACAAAGUU--GCCCCAGGUAAGUUAAUCACUUCCUGGGAAUCCAGUG----CAACGGAG-AUUAUUUAAAACACAACAAAUUUCAUUUGGCCCUACAUAAAU -------------.......(((((((--((((((((.((((.....))))))))))........)----)))).(((-(((...............)))))).))))............. ( -21.46, z-score = -1.71, R) >droWil1.scaffold_180702 3791076 108 - 4511350 ----------CAGCUAUAUACAAUUAU-AUACCUGAUCAGUUGAGUCAUUCAUCGGGUAUUCUUAGUUUCUGGUCUUAAGUCGUUUACAAAUUAUUACAGGGUACCUCAGCAUUUGGAU-- ----------(((.((((((....)))-))).)))....((((((..((((((((((.((.....)).)))))).....((.....))...........))))..))))))........-- ( -17.30, z-score = 0.35, R) >droVir3.scaffold_12970 4425622 115 + 11907090 AUGAUGACUGAAGACAAUAACAAGACUGGUACUCGACCACAUAAAAGUUAUCUCUAGCAUCAAGAG------AUCCUAUAUCUCUUAGAGAACCGGGCAUCAAAUAUAGACUCUCGUACAG .(((((.((.............)).(((((.(((............((((....))))...(((((------((.....))))))).))).))))).)))))................... ( -21.72, z-score = -0.42, R) >consensus _____________CAAAGAACAAAGUU__UACCCAGGUAAGUUAAUCACUUCCUGGGGAUCCAGAG____CAACGAAGAAUUAUUUAAAACACAACAAAUUUCAUUUAGCCCUACAUAAAU ...............................((((((.(((.......)))))))))................................................................ ( -7.39 = -7.03 + -0.36)


| Location | 10,535,502 – 10,535,594 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 57.95 |
| Shannon entropy | 0.77605 |
| G+C content | 0.40355 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.00 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.87 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
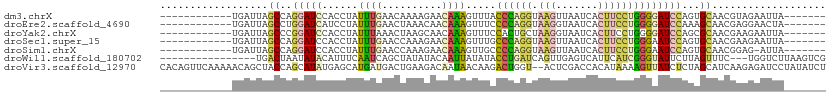
>dm3.chrX 10535502 92 + 22422827 -------UAAUUCUACGUUGCACUGGAUCCCCAGGAAGUGAUUAACUUACCUGGGUAAACUUUGUUCUUUUGUUCAAAUAGGUGGAUCCUGGCUAAUCA------------ -------............((...(((((((((((((((.....)))).)))))......((((..(....)..)))).....))))))..))......------------ ( -25.80, z-score = -1.98, R) >droEre2.scaffold_4690 14158347 92 - 18748788 -------UAGUUCCUCGUUGCUUUGGAUCCCCAGGAAGUGAUUACCUUACCUGGGGAAACUUUGUUGUUUAGUUCAAAUAGGAUGAUCCAGGCUAAUCA------------ -------(((((..(((((..(((((((((((((((((.......))).)))))))((((......)))).)))))))...)))))....)))))....------------ ( -25.00, z-score = -0.81, R) >droYak2.chrX 19104954 92 + 21770863 -------UAAUUCUUCGUUGCGCUGGAUCCCCAGGAAGUGAUUACCUUAGCAGUGGAAACUUUGUUGCUUAGUUUAAAUAGGUGGAUCCGGGCUAAUCA------------ -------............((.(((((((((((((((....)).)))((((((.(....).)))))).............)).))))))))))......------------ ( -27.70, z-score = -1.72, R) >droSec1.super_15 1239054 92 + 1954846 -------UAAUUCUUCGUUGCACUGGAUUCCCAGGAAGUGAUUAACUUACCUGGGCAAACUUUGUUCUUUGGUUCAAAUAGGUGGAUCCUGGCUAAUCA------------ -------............((...(((((((((((((((.....)))).)))))((....((((..(....)..))))...))))))))..))......------------ ( -23.30, z-score = -0.85, R) >droSim1.chrX 8163795 91 + 17042790 -------UAAU-CUCCGUUGCACUGGAUUCCCAGGAAGUGAUUAACUUACCUGGGGCAACUUUGUUCUUUGGUUCAAAUAGGUGGAUCCUGGCUAAUCA------------ -------....-.......((...(((((((((((((((.....)))).))))))((...((((..(....)..))))...)).)))))..))......------------ ( -24.80, z-score = -0.74, R) >droWil1.scaffold_180702 3791112 92 + 4511350 CGACUUAAGACCA---GAAACUAAGAAUACCCGAUGAAUGACUCAACUGAUCAGGUAUAUAAUUGUAUAUAGCUGAUUGAAAUGUAUAUUAGUCA---------------- .............---...(((((..((((....(((.....)))...((((((.((((((....)))))).)))))).....)))).)))))..---------------- ( -15.90, z-score = -1.27, R) >droVir3.scaffold_12970 4425660 109 - 11907090 AGAUAUAGGAUCUCUUGAUGCUAGAGAUAACUUUUAUGUGGUCGAGU--ACCAGUCUUGUUAUUGUCUUCAGUCAUCAUGCUCAUAUGCUGGUAGCUGUUUUUGAACUGUG ((((.(((.(((....))).)))..((((((.......((((.....--)))).....)))))))))).(((..((((.((......))))))..)))............. ( -21.00, z-score = 1.33, R) >consensus _______UAAUUCUUCGUUGCACUGGAUCCCCAGGAAGUGAUUAACUUACCUGGGGAAACUUUGUUCUUUAGUUCAAAUAGGUGGAUCCUGGCUAAUCA____________ ...................((.(((((((((((((((((.....)))).)))))).....((((..........))))......))))))))).................. (-11.15 = -11.00 + -0.15)


| Location | 10,535,502 – 10,535,594 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 57.95 |
| Shannon entropy | 0.77605 |
| G+C content | 0.40355 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -8.42 |
| Energy contribution | -8.39 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10535502 92 - 22422827 ------------UGAUUAGCCAGGAUCCACCUAUUUGAACAAAAGAACAAAGUUUACCCAGGUAAGUUAAUCACUUCCUGGGGAUCCAGUGCAACGUAGAAUUA------- ------------......(((.((((((.......(((((...........))))).(((((.((((.....))))))))))))))).).))............------- ( -24.50, z-score = -2.45, R) >droEre2.scaffold_4690 14158347 92 + 18748788 ------------UGAUUAGCCUGGAUCAUCCUAUUUGAACUAAACAACAAAGUUUCCCCAGGUAAGGUAAUCACUUCCUGGGGAUCCAAAGCAACGAGGAACUA------- ------------(((((......)))))((((.((((..........))))(..((((((((.(((.......)))))))))))..).........))))....------- ( -25.70, z-score = -1.89, R) >droYak2.chrX 19104954 92 - 21770863 ------------UGAUUAGCCCGGAUCCACCUAUUUAAACUAAGCAACAAAGUUUCCACUGCUAAGGUAAUCACUUCCUGGGGAUCCAGCGCAACGAAGAAUUA------- ------------......(((.((((((.((...........((((.............)))).(((.........))))))))))).).))............------- ( -19.12, z-score = -0.64, R) >droSec1.super_15 1239054 92 - 1954846 ------------UGAUUAGCCAGGAUCCACCUAUUUGAACCAAAGAACAAAGUUUGCCCAGGUAAGUUAAUCACUUCCUGGGAAUCCAGUGCAACGAAGAAUUA------- ------------......(((.((((..................((((...)))).((((((.((((.....)))))))))).)))).).))............------- ( -18.70, z-score = -0.67, R) >droSim1.chrX 8163795 91 - 17042790 ------------UGAUUAGCCAGGAUCCACCUAUUUGAACCAAAGAACAAAGUUGCCCCAGGUAAGUUAAUCACUUCCUGGGAAUCCAGUGCAACGGAG-AUUA------- ------------(((((..(((((.....)))...................(((((((((((.((((.....))))))))))........))))))).)-))))------- ( -22.10, z-score = -1.22, R) >droWil1.scaffold_180702 3791112 92 - 4511350 ----------------UGACUAAUAUACAUUUCAAUCAGCUAUAUACAAUUAUAUACCUGAUCAGUUGAGUCAUUCAUCGGGUAUUCUUAGUUUC---UGGUCUUAAGUCG ----------------.((((.............(((((.((((((....)))))).)))))....(((((((...((..((....))..))...---))).)))))))). ( -15.20, z-score = -0.81, R) >droVir3.scaffold_12970 4425660 109 + 11907090 CACAGUUCAAAAACAGCUACCAGCAUAUGAGCAUGAUGACUGAAGACAAUAACAAGACUGGU--ACUCGACCACAUAAAAGUUAUCUCUAGCAUCAAGAGAUCCUAUAUCU ....(((((......((.....))...))))).(((((.(((.(((...((((.....((((--.....)))).......))))))).))))))))............... ( -16.60, z-score = -0.02, R) >consensus ____________UGAUUAGCCAGGAUCCACCUAUUUGAACUAAAGAACAAAGUUUCCCCAGGUAAGUUAAUCACUUCCUGGGGAUCCAGAGCAACGAAGAAUUA_______ ..................((..(((((......((((..........)))).....((((((.(((.......))))))))))))))...))................... ( -8.42 = -8.39 + -0.03)
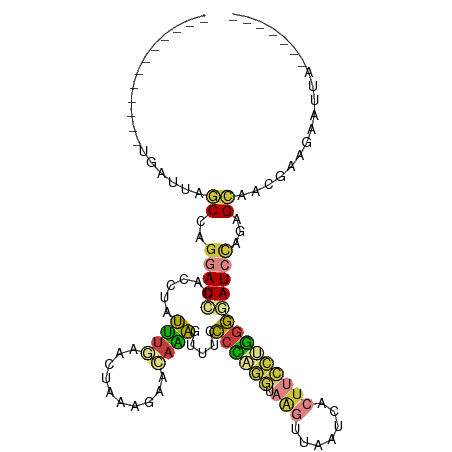

| Location | 10,535,508 – 10,535,599 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 58.40 |
| Shannon entropy | 0.75563 |
| G+C content | 0.42260 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.00 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.87 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10535508 91 + 22422827 -----UACGUUGCACUGGAUCCCCAGGAAGUGAUUAACUUACCUGGGUAAACUUUGUUCUUUUGUUCAAAUAGGUGGAUCCUGGCUAAUCAUGGCA--------------- -----.........(.(((((((((((((((.....)))).)))))......((((..(....)..)))).....)))))).)((((....)))).--------------- ( -26.00, z-score = -1.35, R) >droEre2.scaffold_4690 14158353 91 - 18748788 -----CUCGUUGCUUUGGAUCCCCAGGAAGUGAUUACCUUACCUGGGGAAACUUUGUUGUUUAGUUCAAAUAGGAUGAUCCAGGCUAAUCAUGGCA--------------- -----.(((((..(((((((((((((((((.......))).)))))))((((......)))).)))))))...))))).....((((....)))).--------------- ( -25.80, z-score = -0.86, R) >droYak2.chrX 19104960 91 + 21770863 -----UUCGUUGCGCUGGAUCCCCAGGAAGUGAUUACCUUAGCAGUGGAAACUUUGUUGCUUAGUUUAAAUAGGUGGAUCCGGGCUAAUCAUGGCA--------------- -----.........(((((((((((((((....)).)))((((((.(....).)))))).............)).))))))))((((....)))).--------------- ( -28.00, z-score = -1.26, R) >droSec1.super_15 1239060 91 + 1954846 -----UUCGUUGCACUGGAUUCCCAGGAAGUGAUUAACUUACCUGGGCAAACUUUGUUCUUUGGUUCAAAUAGGUGGAUCCUGGCUAAUCAUGGCA--------------- -----.........(.(((((((((((((((.....)))).)))))((....((((..(....)..))))...)))))))).)((((....)))).--------------- ( -23.50, z-score = -0.29, R) >droSim1.chrX 8163800 91 + 17042790 -----UCCGUUGCACUGGAUUCCCAGGAAGUGAUUAACUUACCUGGGGCAACUUUGUUCUUUGGUUCAAAUAGGUGGAUCCUGGCUAAUCAUGGCA--------------- -----.((((.((...(((((((((((((((.....)))).))))))((...((((..(....)..))))...)).)))))..)).....))))..--------------- ( -26.20, z-score = -0.57, R) >droWil1.scaffold_180702 3791120 89 + 4511350 ---GACCAGAAACUAAGAAUACCCGAUGAAUGACUCAACUGAUCAGGUAUAUAAUUGUAUAUAGCUGAUUGAAAUGUAUAUUAGUCAUAAGA------------------- ---........(((((..((((....(((.....)))...((((((.((((((....)))))).)))))).....)))).))))).......------------------- ( -15.90, z-score = -1.55, R) >droVir3.scaffold_12970 4425668 109 - 11907090 GAUCUCUUGAUGCUAGAGAUAACUUUUAUGUGGUCGAGU--ACCAGUCUUGUUAUUGUCUUCAGUCAUCAUGCUCAUAUGCUGGUAGCUGUUUUUGAACUGUGUGCUGGCA .((((((.......))))))............(((.(((--((((((....(((..(....(((..((((.((......))))))..))).)..))))))).)))))))). ( -26.10, z-score = 0.12, R) >consensus _____UUCGUUGCACUGGAUCCCCAGGAAGUGAUUAACUUACCUGGGGAAACUUUGUUCUUUAGUUCAAAUAGGUGGAUCCUGGCUAAUCAUGGCA_______________ ...........((.(((((((((((((((((.....)))).)))))).....((((..........))))......))))))))).......................... (-11.15 = -11.00 + -0.15)


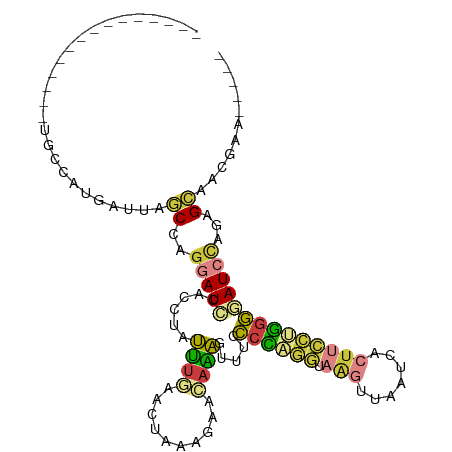
| Location | 10,535,508 – 10,535,599 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 58.40 |
| Shannon entropy | 0.75563 |
| G+C content | 0.42260 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -8.42 |
| Energy contribution | -8.39 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10535508 91 - 22422827 ---------------UGCCAUGAUUAGCCAGGAUCCACCUAUUUGAACAAAAGAACAAAGUUUACCCAGGUAAGUUAAUCACUUCCUGGGGAUCCAGUGCAACGUA----- ---------------...........(((.((((((.......(((((...........))))).(((((.((((.....))))))))))))))).).))......----- ( -24.50, z-score = -2.09, R) >droEre2.scaffold_4690 14158353 91 + 18748788 ---------------UGCCAUGAUUAGCCUGGAUCAUCCUAUUUGAACUAAACAACAAAGUUUCCCCAGGUAAGGUAAUCACUUCCUGGGGAUCCAAAGCAACGAG----- ---------------(((.((((((......))))))......................(..((((((((.(((.......)))))))))))..)...))).....----- ( -25.30, z-score = -2.20, R) >droYak2.chrX 19104960 91 - 21770863 ---------------UGCCAUGAUUAGCCCGGAUCCACCUAUUUAAACUAAGCAACAAAGUUUCCACUGCUAAGGUAAUCACUUCCUGGGGAUCCAGCGCAACGAA----- ---------------...........(((.((((((.((...........((((.............)))).(((.........))))))))))).).))......----- ( -19.12, z-score = -0.40, R) >droSec1.super_15 1239060 91 - 1954846 ---------------UGCCAUGAUUAGCCAGGAUCCACCUAUUUGAACCAAAGAACAAAGUUUGCCCAGGUAAGUUAAUCACUUCCUGGGAAUCCAGUGCAACGAA----- ---------------((((..((((....(((.....)))........................((((((.((((.....))))))))))))))..).))).....----- ( -18.90, z-score = -0.45, R) >droSim1.chrX 8163800 91 - 17042790 ---------------UGCCAUGAUUAGCCAGGAUCCACCUAUUUGAACCAAAGAACAAAGUUGCCCCAGGUAAGUUAAUCACUUCCUGGGAAUCCAGUGCAACGGA----- ---------------..((....((((..(((.....)))..)))).............(((((((((((.((((.....))))))))))........))))))).----- ( -22.10, z-score = -0.92, R) >droWil1.scaffold_180702 3791120 89 - 4511350 -------------------UCUUAUGACUAAUAUACAUUUCAAUCAGCUAUAUACAAUUAUAUACCUGAUCAGUUGAGUCAUUCAUCGGGUAUUCUUAGUUUCUGGUC--- -------------------.((...((((((.((((......(((((.((((((....)))))).)))))...((((........))))))))..))))))...))..--- ( -14.60, z-score = -0.78, R) >droVir3.scaffold_12970 4425668 109 + 11907090 UGCCAGCACACAGUUCAAAAACAGCUACCAGCAUAUGAGCAUGAUGACUGAAGACAAUAACAAGACUGGU--ACUCGACCACAUAAAAGUUAUCUCUAGCAUCAAGAGAUC ............(((((......((.....))...))))).(((((.(((.(((...((((.....((((--.....)))).......))))))).))))))))....... ( -16.60, z-score = 0.40, R) >consensus _______________UGCCAUGAUUAGCCAGGAUCCACCUAUUUGAACUAAAGAACAAAGUUUCCCCAGGUAAGUUAAUCACUUCCUGGGGAUCCAGAGCAACGAA_____ ..........................((..(((((......((((..........)))).....((((((.(((.......))))))))))))))...))........... ( -8.42 = -8.39 + -0.03)
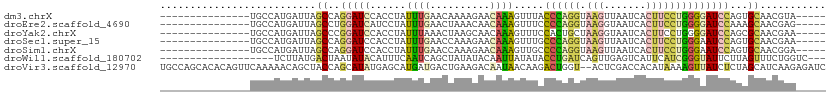
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:51 2011