| Sequence ID | dm3.chr2L |
|---|---|
| Location | 962,114 – 962,221 |
| Length | 107 |
| Max. P | 0.960461 |

| Location | 962,114 – 962,221 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.70 |
| Shannon entropy | 0.62582 |
| G+C content | 0.58272 |
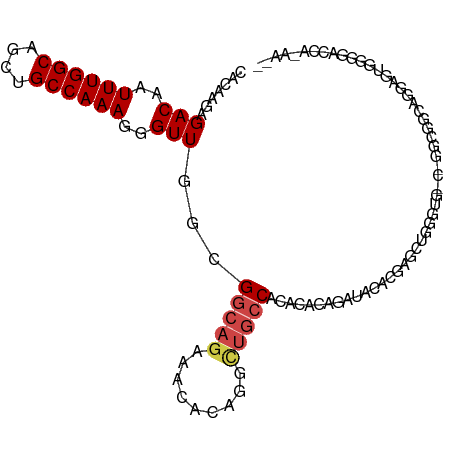
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -15.03 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.56 |
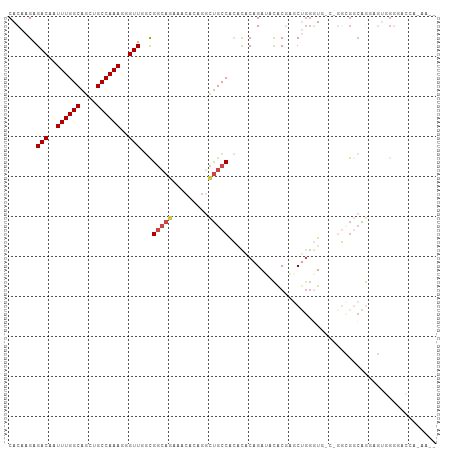
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
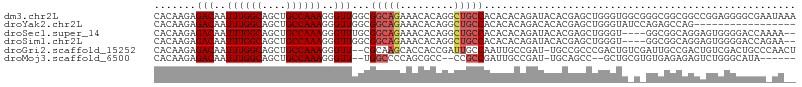
>dm3.chr2L 962114 107 - 23011544 CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUUGGCGGCAGAAACACAGGCUGCCACACACAGAUACACGAGCUGGGUGGCGGGCGGCGGCCGGAGGGGCGAAUAAA .........(..((((((.((((((((....))))))))..........((((((.(((.(((.........))).)))...)))))).))))))..)......... ( -40.00, z-score = -2.96, R) >droYak2.chr2L 950374 90 - 22324452 CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUUGGCGGCAGAAACACAGGCUGCCACACACAGACACACGAGCUGGGUAUCCAGAGCCAG----------------- .......(((..((((((....))))))..)))((((((((.........))))).................((((....)))).)))..----------------- ( -28.90, z-score = -2.06, R) >droSec1.super_14 933043 101 - 2068291 CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUUUGCGGCAGAAACACAGGCUGCCACACACAGAUACACGAGCUGGGU----GGCGGCAGGAGUGGGGACCAAAA-- ......((((..((((((....))))))..))))..(((....((.(..((((((((...(((.........))).))----))))))..).))...).))....-- ( -34.40, z-score = -2.34, R) >droSim1.chr2L 954221 101 - 22036055 CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUUGGCGGCAGAAACACAGGCUGCCACACACAGAUACACGAGCUGGGU----GGCGGCAGGAGUGGGGACCAGAA-- ............((((((.((((((((....)))))))).....(((..((((((((...(((.........))).))----))))))....)))..).))))).-- ( -38.50, z-score = -3.63, R) >droGri2.scaffold_15252 7397346 104 - 17193109 CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUU--CGCAAGCACCACCGAUUGCCAAUUGCCGAU-UGCCGCCCGACUGUCGAUUGCCGACUGUCGACUGCCCAACU .......(.(((((.(((((.((.(((..(((.--(....).))).....))).)).))))))))-)).)((.((((.((((.....)))).))))...))...... ( -28.90, z-score = -0.77, R) >droMoj3.scaffold_6500 8695018 94 + 32352404 CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUU--UGGCCCCAGCGCC--CCGCCGAUUGCCGAU-UGCAGCC--GCUGCGUGUGAGAGAGUCUGGGCAUA------ ......((((..((((((....))))))..)))--).((....))(((--(.(((..(..(((..-.((....--))..)).)..)..).))..))))...------ ( -28.00, z-score = 1.37, R) >consensus CACAAGAGACAAUUUGGCAGCUGCCAAAGGGUUGGCGGCAGAAACACAGGCUGCCACACACAGAUACACGAGCUGGGUG_C_GGCGGCAGGAGUGGGGACCA_AA__ .......(((..((((((....))))))..)))...(((((.........))))).................................................... (-15.03 = -15.58 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:24 2011