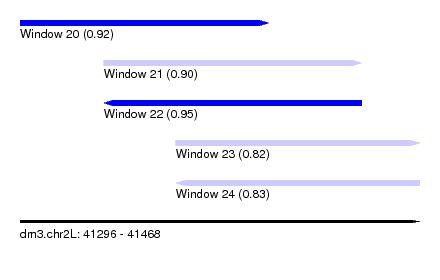
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 41,296 – 41,468 |
| Length | 172 |
| Max. P | 0.951734 |

| Location | 41,296 – 41,403 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.13 |
| Shannon entropy | 0.55987 |
| G+C content | 0.45139 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -13.71 |
| Energy contribution | -13.79 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
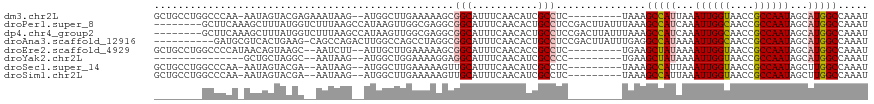
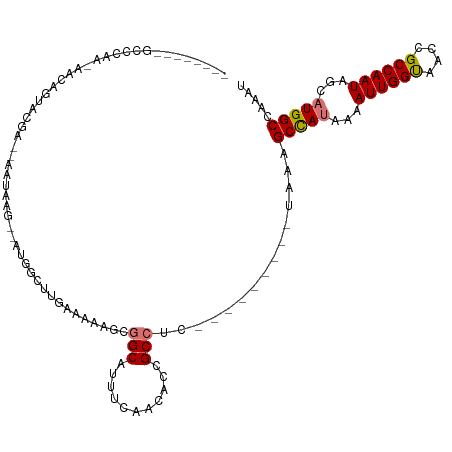
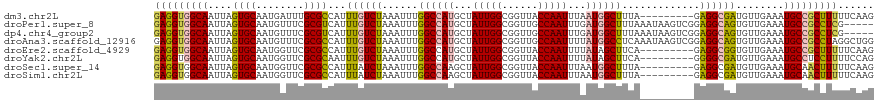
>dm3.chr2L 41296 107 + 23011544 GCUGCCUGGCCCAA-AAUAGUACGAGAAAUAAG--AUGGCUUGAAAAAGCGGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCAUGGCCAAAU ......(((((...-.................(--(((((((.....((.(((...........))).)---------).))))))))..((((((....)))))).....)))))... ( -27.90, z-score = -1.35, R) >droPer1.super_8 3791962 111 + 3966273 --------GCUUCAAAGCUUUAUGGUCUUUAAGCCAUAAGUUGGCGAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAU --------((((.((((((....)).)))))))).(((((((((..((((((..........))))))))))))))).....(((((...((((((....))))))...)))))..... ( -36.80, z-score = -3.35, R) >dp4.chr4_group2 1132589 111 - 1235136 --------GCUUCAAAGCUUUAUGGUCUUUAAGCCAUAAGUUGGCGAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAU --------((((.((((((....)).)))))))).(((((((((..((((((..........))))))))))))))).....(((((...((((((....))))))...)))))..... ( -36.80, z-score = -3.35, R) >droAna3.scaffold_12916 2027000 108 + 16180835 ----------GAUGCGUCACUGAAG-CAGCCAGACUUGGCCAGCCUAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUGAGGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAU ----------((((.(((......(-(.(((......)))..))...((.((((.........)))).))))).))))...((((((...((((((....))))))...)))))).... ( -35.60, z-score = -1.95, R) >droEre2.scaffold_4929 67103 106 + 26641161 GCUGCCUGGCCCCAUAACAGUAAGC--AAUCUU--AUUGCUUGAAAAAGCGGCAUUUCAACACCGCCUC---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAU ......(((((......((((((((--(((...--)))))))).....((((..........))))..)---------))..(((((.....((((....)))))))))..)))))... ( -28.50, z-score = -1.61, R) >droYak2.chr2L 34859 91 + 22324452 ---------------GCUGCUAGGC--AAUAAG--AUGGCUGGAAAAGGAGGCAUUUCAACAUCGCCCC---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAU ---------------...((((.((--......--((((((.....(((.(((...........)))))---------)..))))))...((((((....)))))).)).))))..... ( -25.90, z-score = -0.77, R) >droSec1.super_14 45979 105 + 2068291 GCUGCCUGGCCCAA-AAUAGUACGA--AAUAAG--AUGGCUUGAAAAAGUUGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCUUGGCCAAAU ......(((((...-..........--.....(--(((((((.....((..((...........))..)---------).))))))))..((((((....)))))).....)))))... ( -24.40, z-score = -0.96, R) >droSim1.chr2L 46578 105 + 22036055 GCUGCCUGGCCCAA-AAUAGUACGA--AAUAAG--AUGGCUUGAAAAAGUUGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCUUGGCCAAAU ......(((((...-..........--.....(--(((((((.....((..((...........))..)---------).))))))))..((((((....)))))).....)))))... ( -24.40, z-score = -0.96, R) >consensus ________GCCCAA_AACAGUACGA__AAUAAG__AUGGCUUGAAAAAGCGGCAUUUCAACACCGCCUC_________UAAAGCCAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAU ..................................................(((...........)))...............(((((...((((((....))))))...)))))..... (-13.71 = -13.79 + 0.08)
| Location | 41,332 – 41,443 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.26811 |
| G+C content | 0.44478 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
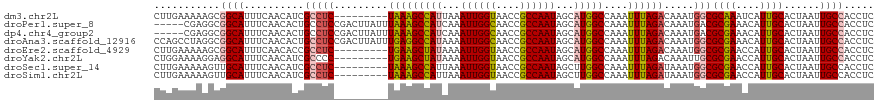
>dm3.chr2L 41332 111 + 23011544 CUUGAAAAAGCGGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCAAAUCAUUGCACUAAUUGCCACCUC (((....))).((((..........(((((---------(((((((((...((((((....))))))...)))))....)))))).....)))((((....))))......))))..... ( -29.10, z-score = -2.68, R) >droPer1.super_8 3791998 115 + 3966273 -----CGAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUUGCACUAAUUGCCACCUC -----.((((.((((......................(((((((((((...((((((....))))))...)))))....))))))........((((....))))......)))).)))) ( -30.20, z-score = -2.94, R) >dp4.chr4_group2 1132625 115 - 1235136 -----CGAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUUGCACUAAUUGCCACCUC -----.((((.((((......................(((((((((((...((((((....))))))...)))))....))))))........((((....))))......)))).)))) ( -30.20, z-score = -2.94, R) >droAna3.scaffold_12916 2027028 120 + 16180835 CCAGCCUAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUGAGGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAAACAUUGCACUAAUUGCCACCUC .......(((.((((..........(((......(((..(..((((((...((((((....))))))...))))))..)..)))......)))((((....))))......)))).))). ( -35.40, z-score = -2.50, R) >droEre2.scaffold_4929 67138 111 + 26641161 CUUGAAAAAGCGGCAUUUCAACACCGCCUC---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUC (((....))).((((..........(((((---------(((((((((...((((((....))))))...)))))....)))))).....)))((((....))))......))))..... ( -26.60, z-score = -1.55, R) >droYak2.chr2L 34879 111 + 22324452 CUGGAAAAGGAGGCAUUUCAACAUCGCCCC---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUUGCGCGAACCAUUGCACUAAUUGCCACCUC .......(((.((((..............(---------(((((((((...((((((....))))))...)))))....))))).........((((....))))......)))).))). ( -23.60, z-score = -0.22, R) >droSec1.super_14 46013 111 + 2068291 CUUGAAAAAGUUGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCUUGGCCAAAUUUAGAUAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUC .........((.(((..........(((((---------((((((((....((((((....))))))....))))....)))))).....)))((((....))))......))).))... ( -24.20, z-score = -1.36, R) >droSim1.chr2L 46612 111 + 22036055 CUUGAAAAAGUUGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCUUGGCCAAAUUUAGAUAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUC .........((.(((..........(((((---------((((((((....((((((....))))))....))))....)))))).....)))((((....))))......))).))... ( -24.20, z-score = -1.36, R) >consensus CUUGAAAAAGCGGCAUUUCAACACCGCCUC_________UAAAGCCAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUC ...........((((..........(((...............(((((...((((((....))))))...)))))...............)))((((....))))......))))..... (-21.83 = -22.17 + 0.35)
| Location | 41,332 – 41,443 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.26811 |
| G+C content | 0.44478 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -29.96 |
| Energy contribution | -29.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
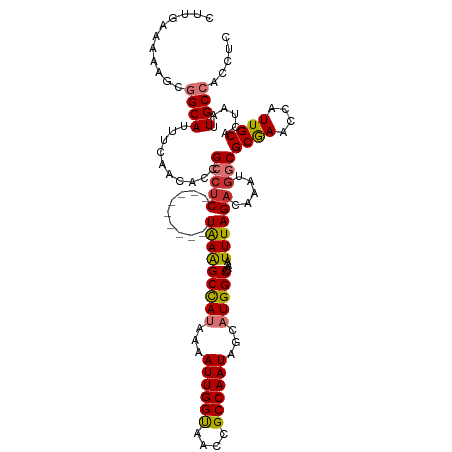
>dm3.chr2L 41332 111 - 23011544 GAGGUGGCAAUUAGUGCAAUGAUUUGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUAAUGGCUUUA---------GAGGCGAUGUUGAAAUGCCGCUUUUUCAAG (((((((((....((((((....))))))((.((((((...(..((((((...(((((......)))))...))))))..)---------.))))))...))...)))))))))...... ( -39.20, z-score = -3.16, R) >droPer1.super_8 3791998 115 - 3966273 GAGGUGGCAAUUAGUGCAAUGUUUCGCGUCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUGAUGGCUUUAAAUAAGUCGGAGGCAGUGUUGAAAUGCCGCCUCG----- (((((((((.((...(((.((((((.((..((((((.(((....((((((...((((((....))))))...))))))))).)))))))))))))).)))..)).))))))))).----- ( -45.90, z-score = -4.35, R) >dp4.chr4_group2 1132625 115 + 1235136 GAGGUGGCAAUUAGUGCAAUGUUUCGCGUCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUGAUGGCUUUAAAUAAGUCGGAGGCAGUGUUGAAAUGCCGCCUCG----- (((((((((.((...(((.((((((.((..((((((.(((....((((((...((((((....))))))...))))))))).)))))))))))))).)))..)).))))))))).----- ( -45.90, z-score = -4.35, R) >droAna3.scaffold_12916 2027028 120 - 16180835 GAGGUGGCAAUUAGUGCAAUGUUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUUAUGGCCUCAAAUAAGUCGGAGGCAGUGUUGAAAUGCCGCCUAGGCUGG .((((((((.((((..(...(.....)(((.((((.((......(((((((..((((((....))))))..))))))).......)).))))))).)..))))..))))))))....... ( -45.42, z-score = -2.50, R) >droEre2.scaffold_4929 67138 111 - 26641161 GAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUAGCUUCA---------GAGGCGGUGUUGAAAUGCCGCUUUUUCAAG ..((((((..((((.((((((((....)))).)))))))).....((((......)))).))))))..............(---------((((((((((....)))))))))))..... ( -34.40, z-score = -1.37, R) >droYak2.chr2L 34879 111 - 22324452 GAGGUGGCAAUUAGUGCAAUGGUUCGCGCAAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUAGCUUCA---------GGGGCGAUGUUGAAAUGCCUCCUUUUCCAG ((((.((((....((((........))))...((((((......(((.(((..(((((......)))))..))).)))...---------.))))))........)))).))))...... ( -30.50, z-score = -0.19, R) >droSec1.super_14 46013 111 - 2068291 GAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUAUCUAAAUUUGGCCAAGCUAUUGGCGGUUACCAAUUUAAUGGCUUUA---------GAGGCGAUGUUGAAAUGCAACUUUUUCAAG ..((((((..((((...((((((....))))))...)))).....(((((....))))).))))))......((.(((...---------..))).)).((((((........)))))). ( -29.00, z-score = -0.55, R) >droSim1.chr2L 46612 111 - 22036055 GAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUAUCUAAAUUUGGCCAAGCUAUUGGCGGUUACCAAUUUAAUGGCUUUA---------GAGGCGAUGUUGAAAUGCAACUUUUUCAAG ..((((((..((((...((((((....))))))...)))).....(((((....))))).))))))......((.(((...---------..))).)).((((((........)))))). ( -29.00, z-score = -0.55, R) >consensus GAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUAAUGGCUUUA_________GAGGCGAUGUUGAAAUGCCGCCUUUUCAAG (((((((((....((((........))))...((((((......((((((...(((((......)))))...)))))).............))))))........)))))))))...... (-29.96 = -29.76 + -0.20)
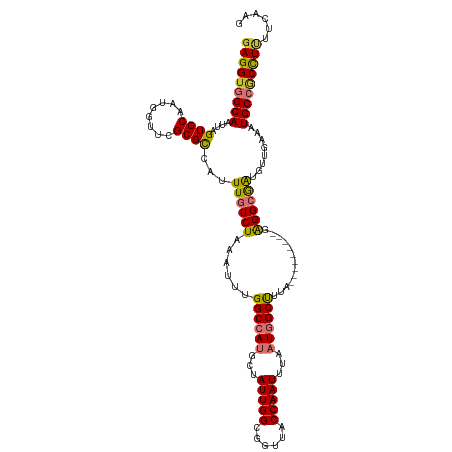
| Location | 41,363 – 41,468 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
| Shannon entropy | 0.36569 |
| G+C content | 0.45529 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -20.04 |
| Energy contribution | -19.94 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
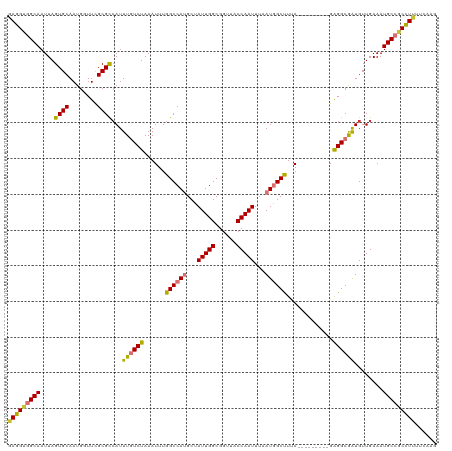
>dm3.chr2L 41363 105 + 23011544 AAAGCCAUUAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCAAAUCAUUGCACUAAUUGCCACCUCGUUUGGCAGCGUCA--AAUGGAUCUUA-------------- ...(((((...((((((....))))))...)))))........(((...(((.((((....)))).))).((((((.......)))))).))).--...........-------------- ( -27.30, z-score = -1.19, R) >droPer1.super_8 3792033 120 + 3966273 AAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUUGCACUAAUUGCCACCUCGUUUGGUGGCAACAU-CGUCGAAUGCAGCCUCGAAAUCCGA ...(((((...((((((....))))))...)))))........(.((..((((((((....)))).....((((((((......))))))))...-.))))..)))....(((.....))) ( -35.70, z-score = -2.61, R) >dp4.chr4_group2 1132660 120 - 1235136 AAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUUGCACUAAUUGCCACCUCGUUUGGUGGCAACAU-CGUCGAAUGCAGCCUCGAAAUCCGA ...(((((...((((((....))))))...)))))........(.((..((((((((....)))).....((((((((......))))))))...-.))))..)))....(((.....))) ( -35.70, z-score = -2.61, R) >droAna3.scaffold_12916 2027068 103 + 16180835 GAGGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAAACAUUGCACUAAUUGCCACCUCGUUUGGCAGC--CG--CAUGGAUCUGA-------------- ..((((((...((((((....))))))...))))))....(((((((..((((((((....))))......(((((.......)))))))--))--..))..)))))-------------- ( -35.10, z-score = -2.41, R) >droEre2.scaffold_4929 67169 114 + 26641161 GAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUCGUUCGGCACCGUCAACCGCUGCCUCGAGAUCUUA------- ...(((((.....((((....))))))))).((((.....((((...(((((......)))))...))))..)))).((((...((((.((.....)).)))).))))......------- ( -30.40, z-score = -1.76, R) >droYak2.chr2L 34910 114 + 22324452 GAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUUGCGCGAACCAUUGCACUAAUUGCCACCUCGUUCGGCAGCGUCAAACGCUGCCUCGAGAUCUUA------- ...(((((.....((((....))))))))).((((..(((((....)))))..((((....)))).......)))).((((...(((((((.....))))))).))))......------- ( -33.80, z-score = -2.74, R) >droSec1.super_14 46044 105 + 2068291 AAAGCCAUUAAAUUGGUAACCGCCAAUAGCUUGGCCAAAUUUAGAUAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUCGUUUGGCAGCGUCA--AAUGGAUCUUA-------------- ...((((....((((((....))))))....)))).......((((...(((((((........)......(((((.......)))))))))))--.....))))..-------------- ( -25.80, z-score = -0.52, R) >droSim1.chr2L 46643 105 + 22036055 AAAGCCAUUAAAUUGGUAACCGCCAAUAGCUUGGCCAAAUUUAGAUAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUCGUUUGGCAGCGUCA--AAUGGAUCUUA-------------- ...((((....((((((....))))))....)))).......((((...(((((((........)......(((((.......)))))))))))--.....))))..-------------- ( -25.80, z-score = -0.52, R) >consensus AAAGCCAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUCGUUUGGCAGCGUCA__CAUGGAUCUGA______________ ...((((......((((((..((((......))))..............(((.((((....)))).))).)))))).......)))).................................. (-20.04 = -19.94 + -0.09)
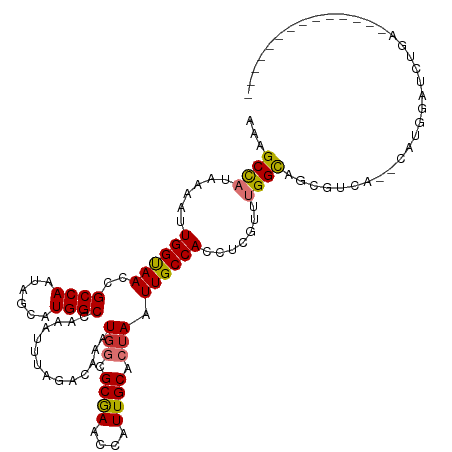
| Location | 41,363 – 41,468 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Shannon entropy | 0.36569 |
| G+C content | 0.45529 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.18 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
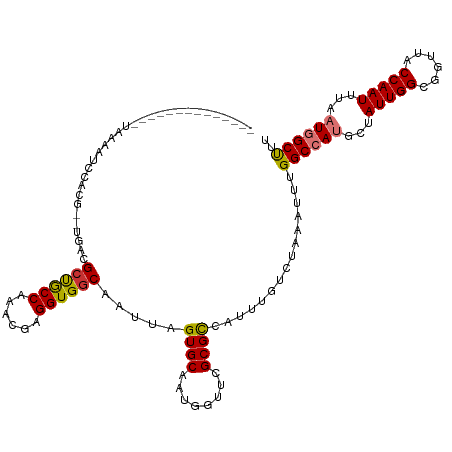
>dm3.chr2L 41363 105 - 23011544 --------------UAAGAUCCAUU--UGACGCUGCCAAACGAGGUGGCAAUUAGUGCAAUGAUUUGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUAAUGGCUUU --------------...........--.(((((..((......))..)).....((((((....)))))).....))).......((((((...(((((......)))))...)))))).. ( -32.20, z-score = -1.79, R) >droPer1.super_8 3792033 120 - 3966273 UCGGAUUUCGAGGCUGCAUUCGACG-AUGUUGCCACCAAACGAGGUGGCAAUUAGUGCAAUGUUUCGCGUCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUGAUGGCUUU ..((((...((.((.(((((..((.-..(((((((((......)))))))))..))..)))))...)).))....))))......((((((...((((((....))))))...)))))).. ( -42.90, z-score = -2.48, R) >dp4.chr4_group2 1132660 120 + 1235136 UCGGAUUUCGAGGCUGCAUUCGACG-AUGUUGCCACCAAACGAGGUGGCAAUUAGUGCAAUGUUUCGCGUCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUGAUGGCUUU ..((((...((.((.(((((..((.-..(((((((((......)))))))))..))..)))))...)).))....))))......((((((...((((((....))))))...)))))).. ( -42.90, z-score = -2.48, R) >droAna3.scaffold_12916 2027068 103 - 16180835 --------------UCAGAUCCAUG--CG--GCUGCCAAACGAGGUGGCAAUUAGUGCAAUGUUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUUAUGGCCUC --------------..((((..(((--..--((..((......))..)).....((((........)))))))..))))......(((((((..((((((....))))))..))))))).. ( -36.40, z-score = -1.81, R) >droEre2.scaffold_4929 67169 114 - 26641161 -------UAAGAUCUCGAGGCAGCGGUUGACGGUGCCGAACGAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUAGCUUC -------((((((.(((.((((.((.....)).))))...)))((((((..((((.((((((((....)))).)))))))).....((((......)))).)))))).))))))....... ( -35.70, z-score = -1.05, R) >droYak2.chr2L 34910 114 - 22324452 -------UAAGAUCUCGAGGCAGCGUUUGACGCUGCCGAACGAGGUGGCAAUUAGUGCAAUGGUUCGCGCAAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUAGCUUC -------....((((((.(((((((.....)))))))...))))))(((((...((((........))))...))))).......(((.(((..(((((......)))))..))).))).. ( -39.00, z-score = -2.36, R) >droSec1.super_14 46044 105 - 2068291 --------------UAAGAUCCAUU--UGACGCUGCCAAACGAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUAUCUAAAUUUGGCCAAGCUAUUGGCGGUUACCAAUUUAAUGGCUUU --------------...((.(((((--.(.((((((((.......))))....)))))))))).))..((((((..........(.(((((....))))).)..........))))))... ( -28.95, z-score = -0.77, R) >droSim1.chr2L 46643 105 - 22036055 --------------UAAGAUCCAUU--UGACGCUGCCAAACGAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUAUCUAAAUUUGGCCAAGCUAUUGGCGGUUACCAAUUUAAUGGCUUU --------------...((.(((((--.(.((((((((.......))))....)))))))))).))..((((((..........(.(((((....))))).)..........))))))... ( -28.95, z-score = -0.77, R) >consensus ______________UAAAAUCCACG__UGACGCUGCCAAACGAGGUGGCAAUUAGUGCAAUGGUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUAAUGGCUUU ...............................((((((......)))))).....((((........))))...............((((((...(((((......)))))...)))))).. (-24.22 = -24.18 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:04:59 2011