| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,465,491 – 10,465,625 |
| Length | 134 |
| Max. P | 0.890020 |

| Location | 10,465,491 – 10,465,625 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 83.95 |
| Shannon entropy | 0.28861 |
| G+C content | 0.52922 |
| Mean single sequence MFE | -47.28 |
| Consensus MFE | -31.43 |
| Energy contribution | -33.03 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
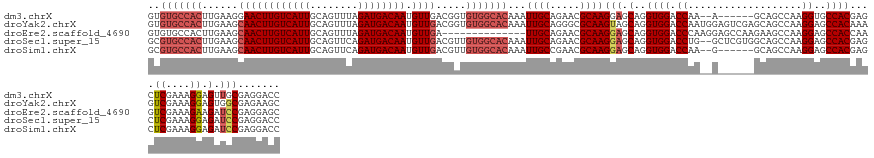
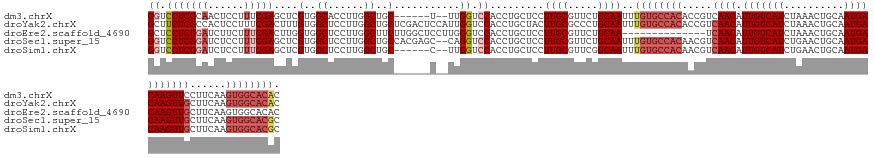
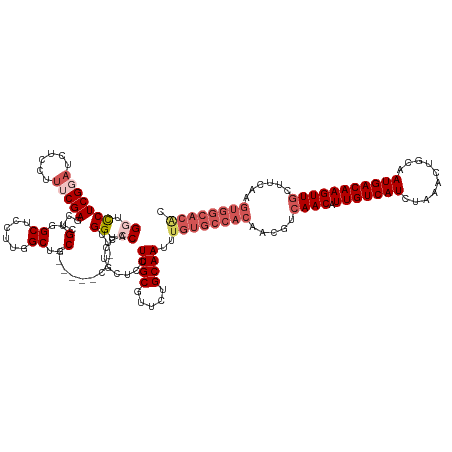
>dm3.chrX 10465491 134 + 22422827 GUGUGCCACUUGAAGGAACUUGUCAUUGCAGUUUAGAUGACAAUGUUGACGGUGUGGCACAAAUUGCAGAACGCAAGGAGCAGGUGGACCAA--A------GCAGCCAAGGUGCCACGAGCUCGAAAGGAGUUGCGAGGACC .((((((((.....(.(((((((((((........)))))))).)))..)...))))))))..((((((..(....)((((..(((((((..--.------........))).))))..))))........))))))..... ( -45.40, z-score = -1.72, R) >droYak2.chrX 19033263 142 + 21770863 GUGUGCCACUUGAAGCAACUUGUCAUUGCAGUUUAGAUGACAAUGUUGACGGUGUGGCACAAAUUGCAGGGCGCAAGUAGCAGGUGGACCAAUGGAGUCGAGCAGCCAAGGAGCCACAAAGUCGAAAGGAGUGGCGAGAAGC .((((((((......((((((((((((........)))))))).)))).....))))))))....((..(((((.....)).(.(.((((....).))).).).))).....(((((....((....)).))))).....)) ( -50.00, z-score = -2.52, R) >droEre2.scaffold_4690 14083582 128 - 18748788 GUGUGCCACUUGAAGCAACUUGUCAUUGCAGUUUAGAUGACAAUGUUGA--------------UUGCAGAACGCAAGGAGCAGGUGGACCCAAGGAGCCAAGAAGCCAAGGAGCCACCAAGUCGAAAGAAGAUCCGAGGAGC ....((..((((..((..(((((..((((((((.(.((....)).).))--------------))))))...)))))..)).(((((..((...(.((......)))..))..)))))...((....)).....))))..)) ( -38.00, z-score = -2.05, R) >droSec1.super_15 1169320 140 + 1954846 GCGUGCCACUUGAAGCAACUUGUCAUUGCAGUUCAGAUGACAAUGUUGACGUUGUGGCACAAAUUGCAGAACGCAAGGAGCAGGUGGACCUG--GCUCGUGGCAGCCAAGGAGCCACGAGCUCGAAAGGAGAUCCGAGGACC ..(((((((......((((((((((((........)))))))).)))).....)))))))...((((.....))))(((.((((....))))--(((((((((..(....).)))))))))((....))...)))....... ( -57.40, z-score = -3.35, R) >droSim1.chrX 8117663 134 + 17042790 GCGUGCCACUUGAAGCAACUUGUCAUUGCAGUUCAGAUGACAAUGUUGACGUUGUGGCACAAAUUGCCGAACGCAAGGAGCAGGUGGACCAA--G------GCAGCCAAGGAGCCACGAGCUCGAAAGGAGAUCCGAGGACC ..(((((((......((((((((((((........)))))))).)))).....)))))))......((...(....)((((..((((.((..--(------(...))..))..))))..))))....((....))..))... ( -45.60, z-score = -1.64, R) >consensus GUGUGCCACUUGAAGCAACUUGUCAUUGCAGUUUAGAUGACAAUGUUGACGGUGUGGCACAAAUUGCAGAACGCAAGGAGCAGGUGGACCAA__G______GCAGCCAAGGAGCCACGAGCUCGAAAGGAGAUCCGAGGACC ..(((((((......((((((((((((........)))))))).)))).....)))))))...((((.....))))(((.(..((((.((...................))..))))....((....)).).)))....... (-31.43 = -33.03 + 1.60)
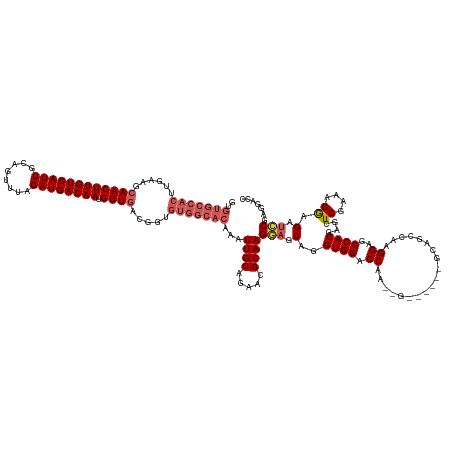
| Location | 10,465,491 – 10,465,625 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 83.95 |
| Shannon entropy | 0.28861 |
| G+C content | 0.52922 |
| Mean single sequence MFE | -42.51 |
| Consensus MFE | -26.88 |
| Energy contribution | -29.36 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.890020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10465491 134 - 22422827 GGUCCUCGCAACUCCUUUCGAGCUCGUGGCACCUUGGCUGC------U--UUGGUCCACCUGCUCCUUGCGUUCUGCAAUUUGUGCCACACCGUCAACAUUGUCAUCUAAACUGCAAUGACAAGUUCCUUCAAGUGGCACAC .((...(((((........((((..((((.(((..((...)------)--..)))))))..)))).)))))....))....((((((((......(((.(((((((..........)))))))))).......)))))))). ( -38.82, z-score = -1.96, R) >droYak2.chrX 19033263 142 - 21770863 GCUUCUCGCCACUCCUUUCGACUUUGUGGCUCCUUGGCUGCUCGACUCCAUUGGUCCACCUGCUACUUGCGCCCUGCAAUUUGUGCCACACCGUCAACAUUGUCAUCUAAACUGCAAUGACAAGUUGCUUCAAGUGGCACAC ((.....(((((.............))))).....(((.....((((.....)))).....((.....)))))..))....((((((((.....((((.(((((((..........)))))))))))......)))))))). ( -40.02, z-score = -2.63, R) >droEre2.scaffold_4690 14083582 128 + 18748788 GCUCCUCGGAUCUUCUUUCGACUUGGUGGCUCCUUGGCUUCUUGGCUCCUUGGGUCCACCUGCUCCUUGCGUUCUGCAA--------------UCAACAUUGUCAUCUAAACUGCAAUGACAAGUUGCUUCAAGUGGCACAC .....(((((......)))))...(((((..((..((((....))))....))..)))))(((..((((......((((--------------(.....(((((((..........))))))))))))..))))..)))... ( -35.10, z-score = -1.48, R) >droSec1.super_15 1169320 140 - 1954846 GGUCCUCGGAUCUCCUUUCGAGCUCGUGGCUCCUUGGCUGCCACGAGC--CAGGUCCACCUGCUCCUUGCGUUCUGCAAUUUGUGCCACAACGUCAACAUUGUCAUCUGAACUGCAAUGACAAGUUGCUUCAAGUGGCACGC ((.(((((((......)))))(((((((((.(....)..)))))))))--..)).))....((...((((.....))))...(((((((.....((((.(((((((.((.....)))))))))))))......))))))))) ( -53.10, z-score = -3.52, R) >droSim1.chrX 8117663 134 - 17042790 GGUCCUCGGAUCUCCUUUCGAGCUCGUGGCUCCUUGGCUGC------C--UUGGUCCACCUGCUCCUUGCGUUCGGCAAUUUGUGCCACAACGUCAACAUUGUCAUCUGAACUGCAAUGACAAGUUGCUUCAAGUGGCACGC ((((....)))).......((((..((((..((..((...)------)--..)).))))..)))).((((.....))))...(((((((.....((((.(((((((.((.....)))))))))))))......))))))).. ( -45.50, z-score = -2.28, R) >consensus GGUCCUCGGAUCUCCUUUCGAGCUCGUGGCUCCUUGGCUGC______C__UUGGUCCACCUGCUCCUUGCGUUCUGCAAUUUGUGCCACAACGUCAACAUUGUCAUCUAAACUGCAAUGACAAGUUGCUUCAAGUGGCACAC ((.(((((((......)))))....(..((......))..)...........)).)).........((((.....))))..((((((((.....((((.(((((((..........)))))))))))......)))))))). (-26.88 = -29.36 + 2.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:42 2011