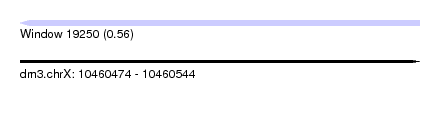
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,460,474 – 10,460,544 |
| Length | 70 |
| Max. P | 0.564412 |

| Location | 10,460,474 – 10,460,544 |
|---|---|
| Length | 70 |
| Sequences | 9 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 59.61 |
| Shannon entropy | 0.85213 |
| G+C content | 0.45740 |
| Mean single sequence MFE | -16.17 |
| Consensus MFE | -6.33 |
| Energy contribution | -6.16 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.55 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10460474 70 - 22422827 -----UAAUUUUGAAUAGUUCUGCGAUAAUGUUUACUACACUGAUGUAUCGAACUGCAGCGGUCUCGAUGACCGU -----..........((((((((((.((.(((.....))).)).))))..))))))..((((((.....)))))) ( -16.60, z-score = -1.53, R) >droSim1.chrX 8110848 70 - 17042790 -----UAAUUUGGAAUAGAUCUGCGAUAAUGUUUACCACACUGAUGUAUCGCACUGCAGCGGUCUCGAUGACCGU -----............(...(((((((....(((......)))..)))))))...).((((((.....)))))) ( -16.50, z-score = -0.74, R) >droSec1.super_15 1158555 70 - 1954846 -----UAAUUUGAAAUAGUUCUGCGAUAAUGUUUACCACACUGAUGUAUCGCACUGCAGCAGUCUCGAUGACCGU -----............((..(((((((....(((......)))..)))))))..)).((.(((.....))).)) ( -12.90, z-score = -0.02, R) >droYak2.chrX 19027708 69 - 21770863 ------UCAUUUGCAUAGUUCUGCGAUAAUGCUUACCACUUUGAUGUAUCGAACCGCAGCGGUCACGAUGACCGU ------.....(((...((((...((((...(..........)...)))))))).)))((((((.....)))))) ( -14.20, z-score = 0.02, R) >droEre2.scaffold_4690 14075918 69 + 18748788 ------CAAUCAGAAUAGUUCUGCGAUAAUGCUUACCACUCUGAUGUAUCGAACUGCAGCGGUCACGAUGACCGU ------..((((((...((...((......))..))...))))))(((......))).((((((.....)))))) ( -16.50, z-score = -0.91, R) >droAna3.scaffold_13335 1090211 70 + 3335858 -----UCCUUUUCGAAGACAGGACCAAUAUGUUGACCAAUAUGAUGGCCCGAACCGCGGCCCUCUCGAUGGCCGU -----...............((.(((.((((((....)))))).))).)).....((((((........)))))) ( -19.40, z-score = -1.21, R) >dp4.chrXL_group1e 5813820 75 - 12523060 UGGCUUUCAUUAGUUUUGUUUGUUGAAGAUGAUCAAAAAUGUGGUGAUGCGUGCUGCUGCCGUCUCAAUGACGGU ..((..((((((.(((((.(..((...))..).)))))...)))))).))........((((((.....)))))) ( -17.80, z-score = -0.61, R) >droPer1.super_21 743971 75 - 1645244 UGGCUUUCAUUAGUUUUGUUUGUUGAAGAUGAUCAAAAAUGUGGUGAUGCGUGCUGCUGCCGUCUCAAUGACGGU ..((..((((((.(((((.(..((...))..).)))))...)))))).))........((((((.....)))))) ( -17.80, z-score = -0.61, R) >droVir3.scaffold_13049 13958570 56 + 25233164 -------------------UACCCAUUGAUCCAUGUUAGACUUUUGGUGCGGUGCGGUGUAGUCGCUGGCGCUGU -------------------...(((..(.((.......)).)..))).(((((((((((....)))).))))))) ( -13.80, z-score = 0.64, R) >consensus _____UAAAUUAGAAUAGUUCUGCGAUAAUGUUUACCACUCUGAUGUAUCGAACUGCAGCGGUCUCGAUGACCGU ..........................................................((((((.....)))))) ( -6.33 = -6.16 + -0.17)

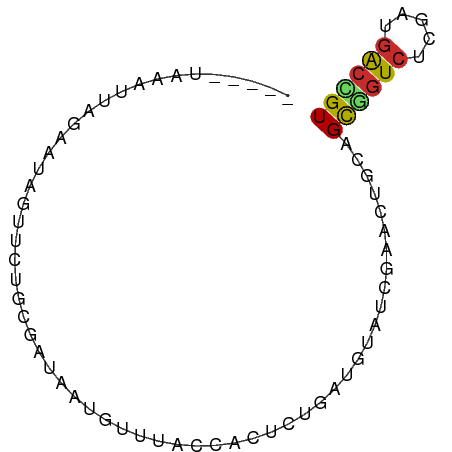

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:40 2011