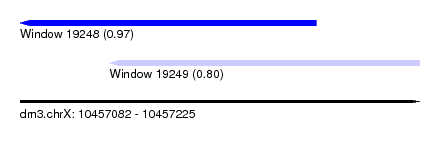
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,457,082 – 10,457,225 |
| Length | 143 |
| Max. P | 0.972804 |

| Location | 10,457,082 – 10,457,188 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Shannon entropy | 0.31496 |
| G+C content | 0.44968 |
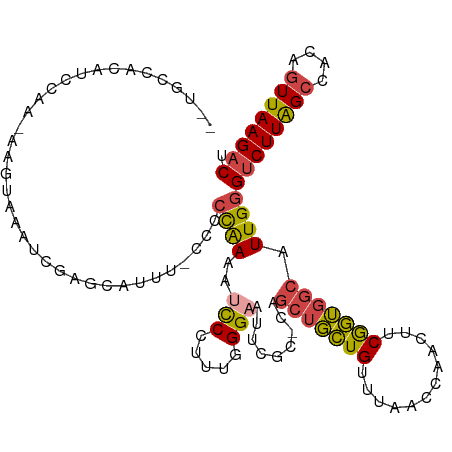
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -21.65 |
| Energy contribution | -20.85 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
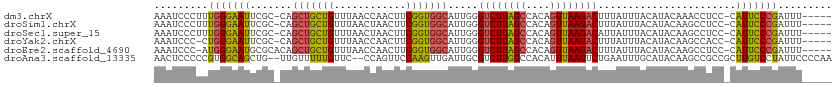
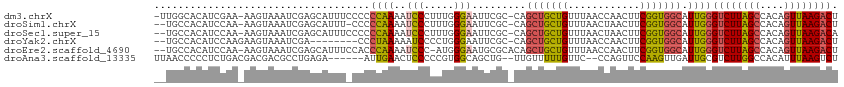
>dm3.chrX 10457082 106 - 22422827 AAAUCCCUUUGGGAAUUCGC-CAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACUUUAUUUACAUACAAACCUCC-CAUUCCCGAUUU----- ........((((((((..((-((.(((..(((.....)))..)))))))...((((((((((....))))))))))...................-.))))))))...----- ( -29.10, z-score = -2.84, R) >droSim1.chrX 8107489 106 - 17042790 AAAUCCCUUUGGGAAUUCGC-CAGCUGCUGUUUAACUAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACUUUAUUUACAUACAAGCCUCC-CAUUCCCGAUUU----- ........((((((((..((-((.(((..(((.....)))..)))))))...((((((((((....))))))))))...................-.))))))))...----- ( -29.80, z-score = -2.72, R) >droSec1.super_15 1155143 106 - 1954846 AAAUCCCUUUGGGAAUUCGC-CAGCUGCUGUUUAACUAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACAUUAUUUACAUACAAGCCUCC-CAUUCCCGAUUU----- ........((((((((..((-((.(((..(((.....)))..))))))).....((((((((....)))))))).....................-.))))))))...----- ( -29.10, z-score = -2.51, R) >droYak2.chrX 19023788 105 - 21770863 AAAUCCC-CUGGGAAUUCGC-CAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACUUUAUUUACAUACAAGCCACC-CAUUCCCGAUUU----- .......-.(((((((..((-.....))...............((((((.((((((((((((....)))))))))...........)))))))))-.)))))))....----- ( -28.90, z-score = -2.30, R) >droEre2.scaffold_4690 14072582 106 + 18748788 AAAUCCC-AUGGGAAUGCGCACAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACUUUAUUUACAUACAAGCCUCC-CAUUCCCGAUUU----- .......-.((((((((.((...((..(((............)))..))...((((((((((....)))))))))).............))....-))))))))....----- ( -30.30, z-score = -2.39, R) >droAna3.scaffold_13335 1086755 109 + 3335858 AACUCCCCCGUGGCAGCUG--UUGUUUUUGUUC--CCAGUUCCAAGUUGAUUGCGUCUUGGCCACAUUUAAGUCUGAAUUUGCAUACAAGCCGCCGCUUGUCCUAUUCCCCAA .........((((((((((--..(.......).--.))))).((((.((....)).)))))))))..........((((......((((((....))))))...))))..... ( -21.20, z-score = -0.63, R) >consensus AAAUCCCUUUGGGAAUUCGC_CAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACUUUAUUUACAUACAAGCCUCC_CAUUCCCGAUUU_____ .........(((((((.......(((((((............))))))).....((((((((....)))))))).......................)))))))......... (-21.65 = -20.85 + -0.80)
| Location | 10,457,114 – 10,457,225 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.44743 |
| G+C content | 0.47321 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
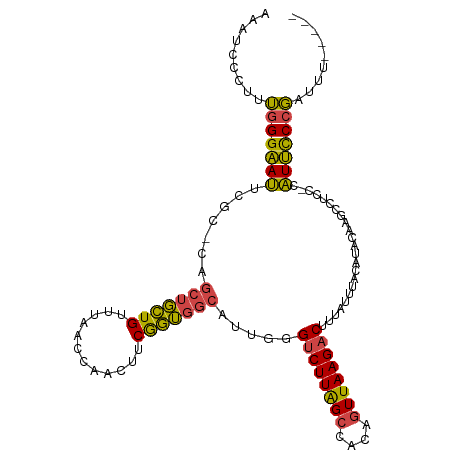
>dm3.chrX 10457114 111 - 22422827 -UUGGCACAUCGAA-AAGUAAAUCGAGCAUUUCCCCCCAAAAUCCCUUUGGGAAUUCGC-CAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACU -.((((...((((.-.......)))).........((((((.....)))))).....))-))((..(((............)))..))....(((((((((....))))))))) ( -33.40, z-score = -2.13, R) >droSim1.chrX 8107521 109 - 17042790 --UGCCACAUCCAA-AAGUAAAUCGAGCAUUU-CCCCCAAAAUCCCUUUGGGAAUUCGC-CAGCUGCUGUUUAACUAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACU --........((((-.................-..((((((.....)))))).....((-((.(((..(((.....)))..))))))).))))((((((((....)))))))). ( -29.80, z-score = -1.76, R) >droSec1.super_15 1155175 110 - 1954846 --UGCCACAUCCAA-AAGUAAAUCGAGCAUUUCCCCCCAAAAUCCCUUUGGGAAUUCGC-CAGCUGCUGUUUAACUAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACA --........((((-....................((((((.....)))))).....((-((.(((..(((.....)))..))))))).))))((((((((....)))))))). ( -30.30, z-score = -1.92, R) >droYak2.chrX 19023820 103 - 21770863 --UGCCACAUCCAAGAAGUAAAUCGA--------CCCUAAAAAUCCCCUGGGAAUUCGC-CAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACU --((((((......(((((.......--------.........(((....)))....((-.....))..........))))).))))))...(((((((((....))))))))) ( -27.30, z-score = -1.28, R) >droEre2.scaffold_4690 14072614 110 + 18748788 --UGCCACAUCCAA-AAGUAAAUCGAGCAUUUCCACCCAAAAUCCC-AUGGGAAUGCGCACAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACU --((((((......-...(((((...(((((((((...........-.)))))))))(((....))).)))))..........))))))...(((((((((....))))))))) ( -32.71, z-score = -2.30, R) >droAna3.scaffold_13335 1086793 104 + 3335858 UUAACCCCCUCUGACGACGACGCCUGAGA------AUUGAACUCCCCCGUGGCAGCUG--UUGUUUUUGUUC--CCAGUUCCAAGUUGAUUGCGUCUUGGCCACAUUUAAGUCU ..................(((....(((.------......)))....((((((((((--..(.......).--.))))).((((.((....)).)))))))))......))). ( -18.50, z-score = 0.60, R) >consensus __UGCCACAUCCAA_AAGUAAAUCGAGCAUUU_CCCCCAAAAUCCCUUUGGGAAUUCGC_CAGCUGCUGUUUAACCAACUUCGGUGGCAUUGGGUCUUAGCCACAGUUAAGACU ....................................((((..(((.....))).........(((((((............))))))).))))((((((((....)))))))). (-17.53 = -17.53 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:39 2011