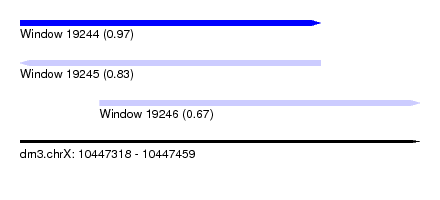
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,447,318 – 10,447,459 |
| Length | 141 |
| Max. P | 0.971403 |

| Location | 10,447,318 – 10,447,424 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.53 |
| Shannon entropy | 0.56193 |
| G+C content | 0.34797 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -10.18 |
| Energy contribution | -9.52 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971403 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
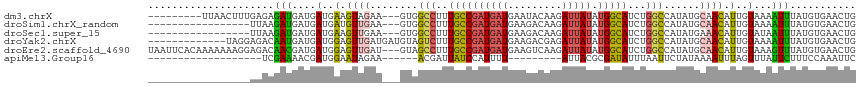
>dm3.chrX 10447318 106 + 22422827 ---------UUAACUUUGAGAGAUGAUGAUGAAGUAGAA---GUGGCCUUUGCCGAUGAUGAAUACAAGAUUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUG ---------...........((((.(..(((..(((...---((((((..((((((((((.........))))).)))))...)))))).)))..)))..)...)))).......... ( -25.80, z-score = -2.74, R) >droSim1.chrX_random 2969025 98 + 5698898 -----------------UUAAGAUGAUGAUGAUGUUGAA---GUGGCCUUUGCCGAUGAUGAAGACAAGAUUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUG -----------------...((((....(..((((((..---((((((..((((((((((.........))))).)))))...))))))...))))))..)...)))).......... ( -29.00, z-score = -4.18, R) >droSec1.super_15 1145468 98 + 1954846 -----------------UUAAGAUGAUGAUGAAGUUGAA---GUGGCCUUUGCCGAUGAUGAAGACAAGAUUAUAUGGCAUCUGGCCAUAUGAAACAUUGUAUAAUUUAUGUGAACUG -----------------...((((.(..(((...((.(.---((((((..((((((((((.........))))).)))))...)))))).).)).)))..)...)))).......... ( -23.00, z-score = -2.73, R) >droYak2.chrX 19012334 105 + 21770863 -------------UAGGAGACAAUGAUGAUGGAGUUGAUGAUGUAGUCUUUGCCGAUGAUGAAGACGAGAUUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUG -------------..(....).......(((((....(..((((.((((((.........)))))).....((((((((.....))))))))..))))..)....)))))........ ( -22.90, z-score = -1.14, R) >droEre2.scaffold_4690 14062976 115 - 18748788 UAAUUCACAAAAAAAGGAGACAACGAUGAUGGAGUUGAU---GUAGCCUUUGCCGAUGAUGAAGUCAAGAUUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAGUUUAUGUGAACUG ...((((((...(((((..(((.((((......)))).)---))..)))))..(((((.((....))....((((((((.....))))))))...))))).........))))))... ( -29.00, z-score = -2.17, R) >apiMel3.Group16 3648293 84 + 5631066 -------------------UCGAAAACGAUGGAAUAGAA------ACGAUUAUCCAUUUU---------AUUACGCGAUAUUUAAUUCUAUAAAAUUUAGUUUAUUCUUUCCAAAUUC -------------------(((....)))(((((.((((------..(((((...(((((---------((.....((........)).))))))).)))))..)))))))))..... ( -10.10, z-score = -0.09, R) >consensus _________________AGAAGAUGAUGAUGAAGUUGAA___GUGGCCUUUGCCGAUGAUGAAGACAAGAUUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUG .....................(((....(..(.((((........(((..((((((((((.........))))).)))))...)))......)))).)..)...)))........... (-10.18 = -9.52 + -0.66)

| Location | 10,447,318 – 10,447,424 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.53 |
| Shannon entropy | 0.56193 |
| G+C content | 0.34797 |
| Mean single sequence MFE | -16.27 |
| Consensus MFE | -8.37 |
| Energy contribution | -7.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832348 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 10447318 106 - 22422827 CAGUUCACAUAAAUUUUACAAUGUUGCAUAUGGCCAGAUGCCAUAUAAUCUUGUAUUCAUCAUCGGCAAAGGCCAC---UUCUACUUCAUCAUCAUCUCUCAAAGUUAA--------- ................(((((.(((..(((((((.....)))))))))).))))).........(((....)))..---..............................--------- ( -15.60, z-score = -0.93, R) >droSim1.chrX_random 2969025 98 - 5698898 CAGUUCACAUAAAUUUUACAAUGUUGCAUAUGGCCAGAUGCCAUAUAAUCUUGUCUUCAUCAUCGGCAAAGGCCAC---UUCAACAUCAUCAUCAUCUUAA----------------- ....................((((((.(..(((((.(((........)))(((((.........))))).))))).---.)))))))..............----------------- ( -18.00, z-score = -2.22, R) >droSec1.super_15 1145468 98 - 1954846 CAGUUCACAUAAAUUAUACAAUGUUUCAUAUGGCCAGAUGCCAUAUAAUCUUGUCUUCAUCAUCGGCAAAGGCCAC---UUCAACUUCAUCAUCAUCUUAA----------------- .((((............((((.(((..(((((((.....)))))))))).))))..........(((....)))..---...))))...............----------------- ( -14.20, z-score = -1.67, R) >droYak2.chrX 19012334 105 - 21770863 CAGUUCACAUAAAUUUUACAAUGUUGCAUAUGGCCAGAUGCCAUAUAAUCUCGUCUUCAUCAUCGGCAAAGACUACAUCAUCAACUCCAUCAUCAUUGUCUCCUA------------- .................((((((.((.(((((((.....)))))))......(((((..(.....)..))))).................)).))))))......------------- ( -16.30, z-score = -1.68, R) >droEre2.scaffold_4690 14062976 115 + 18748788 CAGUUCACAUAAACUUUACAAUGUUGCAUAUGGCCAGAUGCCAUAUAAUCUUGACUUCAUCAUCGGCAAAGGCUAC---AUCAACUCCAUCAUCGUUGUCUCCUUUUUUUGUGAAUUA .((((((((.(((....((((((.((.(((((((.....)))))))..................(((....)))..---...........)).))))))......))).)))))))). ( -22.40, z-score = -1.56, R) >apiMel3.Group16 3648293 84 - 5631066 GAAUUUGGAAAGAAUAAACUAAAUUUUAUAGAAUUAAAUAUCGCGUAAU---------AAAAUGGAUAAUCGU------UUCUAUUCCAUCGUUUUCGA------------------- (((..((((((((((.......))))).(((((.....((((.(((...---------...))))))).....------)))))))))).....)))..------------------- ( -11.10, z-score = 0.16, R) >consensus CAGUUCACAUAAAUUUUACAAUGUUGCAUAUGGCCAGAUGCCAUAUAAUCUUGUCUUCAUCAUCGGCAAAGGCCAC___UUCAACUCCAUCAUCAUCGUAA_________________ ..............................(((((...((((......................))))..)))))........................................... ( -8.37 = -7.93 + -0.44)
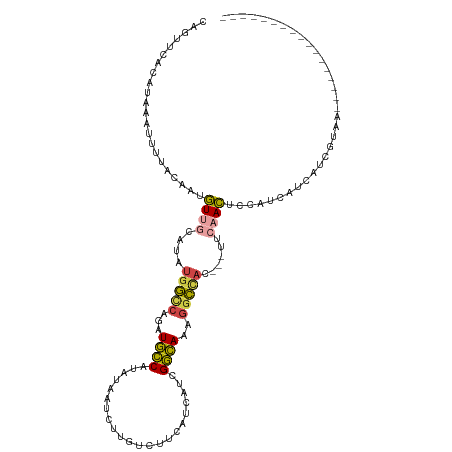
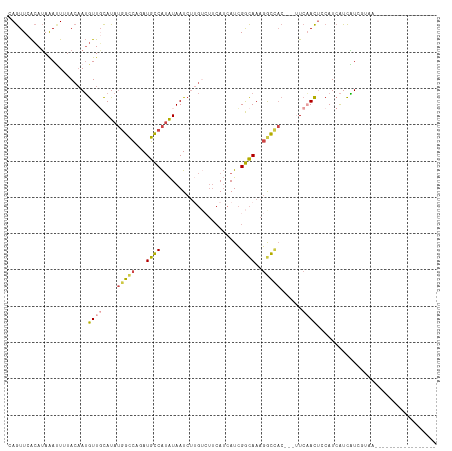
| Location | 10,447,346 – 10,447,459 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Shannon entropy | 0.34309 |
| G+C content | 0.40084 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -19.83 |
| Energy contribution | -22.42 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
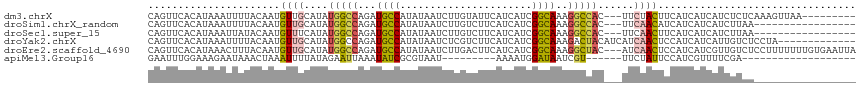
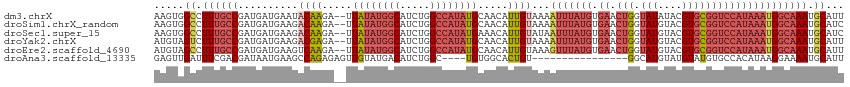
>dm3.chrX 10447346 113 + 22422827 AAGUGGCCUUUGCCGAUGAUGAAUACAAGA--UUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUGGUAUAUACGUGCGGUCCAUAAAUGGCAAAUGCAUU ..((((((..((((((((((.........)--)))).)))))...))))))(((((...((((...(((((((.((.(((.(((....))))))))))))))).)))).))))). ( -33.90, z-score = -2.08, R) >droSim1.chrX_random 2969045 113 + 5698898 AAGUGGCCUUUGCCGAUGAUGAAGACAAGA--UUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUGGUAUGUACGUGCGGUCCAUAAAUGGCAAAUGCAUC ..((((((..((((((((((.........)--)))).)))))...))))))(((((...((((...(((((((.((.(((.(((....))))))))))))))).)))).))))). ( -34.80, z-score = -2.25, R) >droSec1.super_15 1145488 113 + 1954846 AAGUGGCCUUUGCCGAUGAUGAAGACAAGA--UUAUAUGGCAUCUGGCCAUAUGAAACAUUGUAUAAUUUAUGUGAACUGGUAUGUACGUGCGGUCCAUAAAUGGCAAAUGCAUC .....((.((((((..........((((..--(((((((((.....)))))))))....))))...(((((((.((.(((.(((....))))))))))))))))))))).))... ( -33.00, z-score = -2.00, R) >droYak2.chrX 19012361 113 + 21770863 AUGUAGUCUUUGCCGAUGAUGAAGACGAGA--UUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUGGUAUGUACGUGCGGUCCAUAAAUGGCAAAUGCAUU .....((.((((((..........((((..--.((((((((.....)))))))).....))))...(((((((.((.(((.(((....))))))))))))))))))))).))... ( -29.80, z-score = -0.88, R) >droEre2.scaffold_4690 14063013 113 - 18748788 AUGUAGCCUUUGCCGAUGAUGAAGUCAAGA--UUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAGUUUAUGUGAACUGGUAUGUACGUGCGGUCCAUAAAUGGCAAAUGCAUU .....((.((((((.(..(((..((.....--..(((((((.....)))))))))..)))..)...(((((((.((.(((.(((....))))))))))))))))))))).))... ( -31.41, z-score = -0.91, R) >droAna3.scaffold_13335 1076061 95 - 3335858 GAGUUGAUUUCGACGAUAAUGAAGCCAGAGAGUUGUAUGACAUCUGGC----UGUGGCACUGU----------------GGCAUGUAUGUAUGUGCCACAUAAGGAAAAUGCAUU ..((((....))))...((((.(((((((..(((....))).))))))----)(((((((...----------------.(((....)))..)))))))............)))) ( -30.90, z-score = -2.74, R) >consensus AAGUGGCCUUUGCCGAUGAUGAAGACAAGA__UUAUAUGGCAUCUGGCCAUAUGCAACAUUGUAAAAUUUAUGUGAACUGGUAUGUACGUGCGGUCCAUAAAUGGCAAAUGCAUU .....((.((((((..........((((.....((((((((.....)))))))).....))))...(((((((.((....((((....))))..))))))))))))))).))... (-19.83 = -22.42 + 2.59)
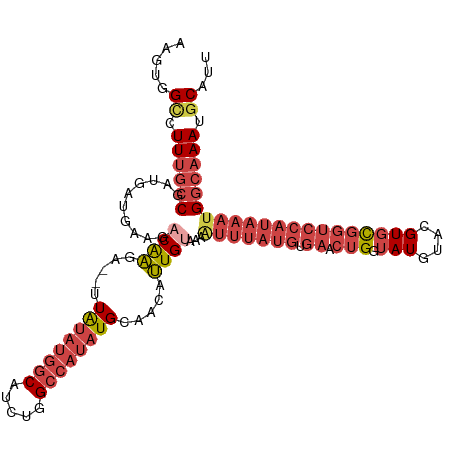
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:36 2011