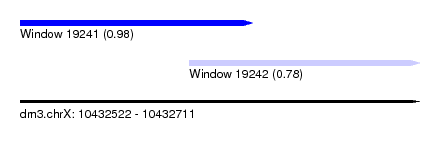
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,432,522 – 10,432,711 |
| Length | 189 |
| Max. P | 0.980173 |

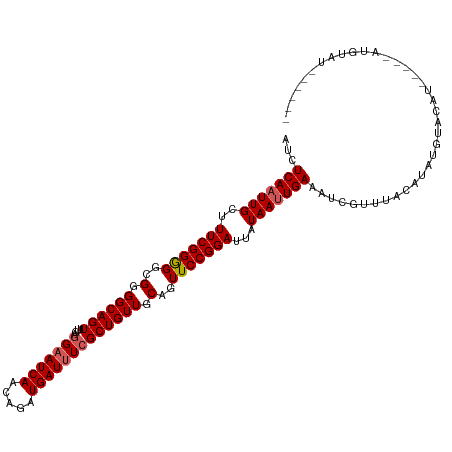
| Location | 10,432,522 – 10,432,632 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.54 |
| Shannon entropy | 0.23407 |
| G+C content | 0.39603 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10432522 110 + 22422827 AUCUCAAUUGCUUUCGGGGGCGUGGCAGUUUGAGGAAUCAACAGAUGAUUUCGCUGUUGCAGUUCCGGAUUAUAAUUGAAAUCGUUUACAUAUGUACAUUCAAUAUGUAU------ ...(((((((...((.((((((..(((((....(((((((.....))))))))))))..).))))).))...))))))).......(((((((.........))))))).------ ( -33.70, z-score = -3.33, R) >droSim1.chrX_random 2950857 105 + 5698898 AUCUCAAUUGCUUUCGGGGGCGGGGCAGUUUGAGGAAUCAACAGAUGAUUUCGCUGUUGCAGUUCCGGAUUAUAAUUGAAAUCGUUUACAUAUUUACAU-----AUGUAU------ ...(((((((...((.((((((.((((((....(((((((.....))))))))))))).).))))).))...))))))).......(((((((....))-----))))).------ ( -30.80, z-score = -3.07, R) >droSec1.super_15 1132237 105 + 1954846 AUCUCAAUUGCUUUCGGGGGCGGGGCAGUUUGAGGAAUCAACAGAUGAUUUCGCUGUUGCAGUUCCGGAUUAUAAUUGAAAUCGUUUACAUAUUUACAU-----AUGUAU------ ...(((((((...((.((((((.((((((....(((((((.....))))))))))))).).))))).))...))))))).......(((((((....))-----))))).------ ( -30.80, z-score = -3.07, R) >droEre2.scaffold_4690 14049274 105 - 18748788 AUCUCAAUUGCUUUCGGAGGCGGGGCAGUUUGAAGAAUCAACAGAUGAUUUCGCUGUUGCAGUUCCGGAUUAUAAAUGAAAUUGUUCACAUAUGUGCAUAUG-CAU---------- ...(((.(((..(((((((..(.((((((..((((.(((....)))..)))))))))).)..)))))))...))).)))..............((((....)-)))---------- ( -22.90, z-score = 0.63, R) >droYak2.chrX 18998780 115 + 21770863 AUCUCAAUUGCUUUCGGAGGCGGGGCAGUUUGAGGCAUCAACAGAUGAUUUCGCUGUUGCGGUUCCGGAUUGUAAUUGAAAUUGUUCACAUGUGUAUGUAUA-UACACAUACAUAC ...((((((((.(((((((.((.((((((..((((((((....)))).)))))))))).)).)))))))..))))))))...........(((((((((...-...))))))))). ( -43.90, z-score = -5.31, R) >consensus AUCUCAAUUGCUUUCGGGGGCGGGGCAGUUUGAGGAAUCAACAGAUGAUUUCGCUGUUGCAGUUCCGGAUUAUAAUUGAAAUCGUUUACAUAUGUACAU_____AUGUAU______ ...(((((((..(((((((..(.((((((....(((((((.....))))))))))))).)..)))))))...)))))))..................................... (-23.52 = -23.88 + 0.36)

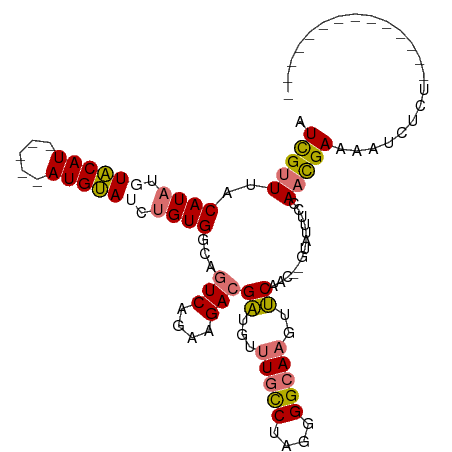
| Location | 10,432,602 – 10,432,711 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.11 |
| Shannon entropy | 0.45911 |
| G+C content | 0.36380 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10432602 109 + 22422827 AUCGUUUACAUAUGUACAUUCAAUAUGUAUCUGUGGCAGUCAGAAGACGAUGUUUGCCUUGGGGCAAGUUCAAC--GUAUUUGCAACGAUGUUCUCUAUUCAAAAAAAUAU ((((((..((((.((((((.....)))))).))))((((((....)))((...(((((....)))))..))...--.....)))))))))..................... ( -25.20, z-score = -1.44, R) >droSim1.chrX_random 2950937 103 + 5698898 AUCGUUUACAUAUUUACAU-----AUGUAUCUGUGGCAGUCAGUAGACGCUGUUUGCCUAGGGGCAAGUGCAAC--GUAUUUCCAACGAUAGUCUCUAUU-GAAGAAUUAU .((.....((((..(((..-----..)))..))))....((((((((.((((((((((....)))))......(--((.......)))))))).))))))-)).))..... ( -25.60, z-score = -1.27, R) >droSec1.super_15 1132317 86 + 1954846 AUCGUUUACAUAUUUACAU-----AUGUAUCUGUGGCAGUCAGUAGAAGAUGUUUGCCUAGGGGCAAGUUCAAC--GUAUUUCCAACGAAUAU------------------ .((((((((((((....))-----))))).....((.(((..((.(((....((((((....))))))))).))--..))).)))))))....------------------ ( -18.20, z-score = -0.99, R) >droEre2.scaffold_4690 14049354 92 - 18748788 AUUGUUCACAUAUGUGCAU-----AUGCAUAAGUGGCAGUCAAAAGACGAUGUAUCUCUAGGGGCAAGUUCAACUGAAAAGUUUAUUGAAAAAAUAU-------------- .((((((.(((.(((((..-----..))))).)))...(((....))).............)))))).(((((((....)))....)))).......-------------- ( -18.20, z-score = -0.86, R) >consensus AUCGUUUACAUAUGUACAU_____AUGUAUCUGUGGCAGUCAGAAGACGAUGUUUGCCUAGGGGCAAGUUCAAC__GUAUUUCCAACGAAAAUCUCU______________ .(((((..((((.((((((.....)))))).))))...(((....)))((...(((((....)))))..)).............)))))...................... (-14.81 = -15.25 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:33 2011