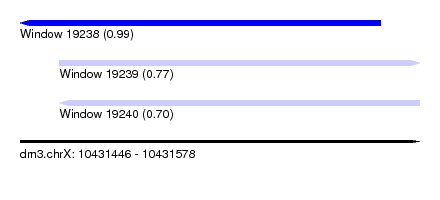
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,431,446 – 10,431,578 |
| Length | 132 |
| Max. P | 0.988430 |

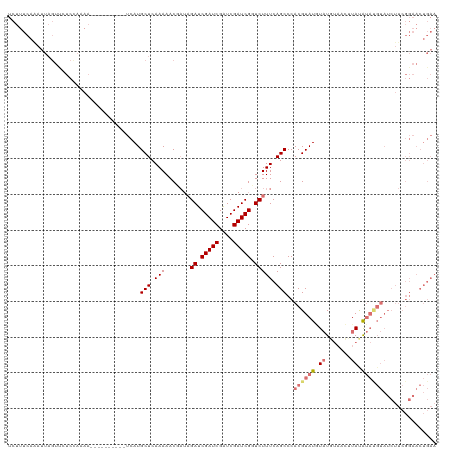
| Location | 10,431,446 – 10,431,565 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.39661 |
| G+C content | 0.38914 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -19.62 |
| Energy contribution | -22.50 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10431446 119 - 22422827 AUACGUAUGUACAUAAGAUAUAUACGUGGGCUGAAAGACCGAUCGAUCGAUCGAUCUAUAGUUUUUGAGCAUGA-UUUCUCGUCUUUGGGAGAACUAAUUUGAAAGAUGUGCGUGCUUAG .(((((((((((....).))))))))))(((((..(((.(((((....))))).))).)))))..((((((((.-.....(((((((.(((.......))).)))))))..)))))))). ( -36.30, z-score = -3.02, R) >droEre2.scaffold_4690 14048245 95 + 18748788 ---------------------AUACGUGGGCUGAAAGACCGAUCGACCGAUCGAUC----GUUUUUGAGCAUGAUUUUCUCGUCUUUUAGAGAACUAAUUUGAAAGACGUGCGUGCUUAA ---------------------.(((((..(((.((((((.((((((....))))))----)))))).)))..........(((((((((((.......))))))))))).)))))..... ( -32.50, z-score = -3.48, R) >droSec1.super_15 1131256 110 - 1954846 AUACAUACAUAAGAUACAUACAUACGUGGGCUGAAAGAACGAUCGAUCGAUCGAUCUAUCGUUUUUGAGCAUGA----------UUUUUGAUGAUGAAUUUGAAAGAUGUGCGUGCUUAG .........((((.(((.(((((......(((..(((((((((.((((....)))).))))))))).)))....----------((((..(........)..))))))))).))))))). ( -27.40, z-score = -1.58, R) >droSim1.chrX_random 2949833 109 - 5698898 AUACAUACAUAAGAUACAUACAUACGUGGGCUGAAAGAACGAUCGAUCGAUCGAUCUAUCGUUUUUGAGCAUGA-----------UUUGGAGAACUAAUCUGAAAGAUGUGCGUGCUUAG .........((((.(((.(((((......(((..(((((((((.((((....)))).))))))))).)))....-----------((..((.......))..))..))))).))))))). ( -28.60, z-score = -2.08, R) >consensus AUACAUACAUAAGAUACAUACAUACGUGGGCUGAAAGAACGAUCGAUCGAUCGAUCUAUCGUUUUUGAGCAUGA__________UUUUGGAGAACUAAUUUGAAAGAUGUGCGUGCUUAG ......................(((((..(((.((((((.((((((....))))))....)))))).)))..........(((((((((((.......))))))))))).)))))..... (-19.62 = -22.50 + 2.88)

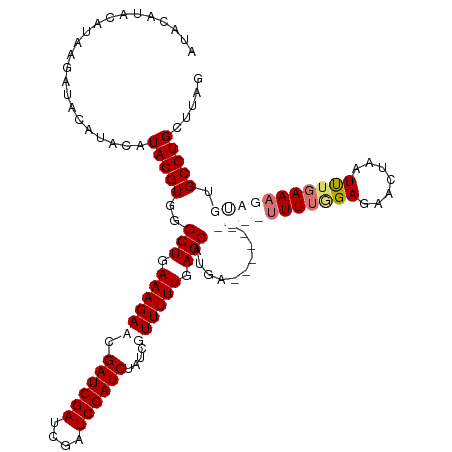
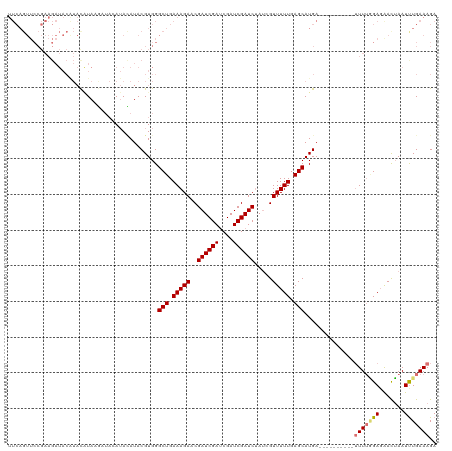
| Location | 10,431,459 – 10,431,578 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.42986 |
| G+C content | 0.37195 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -10.60 |
| Energy contribution | -11.85 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10431459 119 + 22422827 UCUUUCAAAUUAGUUCUCCCAAAGACGAGAAA-UCAUGCUCAAAAACUAUAGAUCGAUCGAUCGAUCGGUCUUUCAGCCCACGUAUAUAUCUUAUGUACAUACGUAUCUUCGGAACUGAA .........((((((((....((((.(((...-.....))).....((..(((((((((....)))))))))...))...((((((.(((.....))).)))))).)))).)))))))). ( -28.00, z-score = -2.54, R) >droEre2.scaffold_4690 14048258 95 - 18748788 UCUUUCAAAUUAGUUCUCUAAAAGACGAGAAAAUCAUGCUCAAAAAC----GAUCGAUCGGUCGAUCGGUCUUUCAGCCCACGUAUCU-UUGAAACUAAA-------------------- ..(((((((...((...((.(((((((((.........))).....(----((((((....))))))))))))).))...)).....)-)))))).....-------------------- ( -22.10, z-score = -2.34, R) >droSec1.super_15 1131269 110 + 1954846 UCUUUCAAAUUCAUCAUCAAAAA----------UCAUGCUCAAAAACGAUAGAUCGAUCGAUCGAUCGUUCUUUCAGCCCACGUAUGUAUGUAUCUUAUGUAUGUAUCUUCGGAACUGAA ................(((....----------....(((.(((((((((.((((....)))).)))))).))).)))..(((((((((.......))))))))).((....))..))). ( -22.20, z-score = -1.51, R) >droSim1.chrX_random 2949846 109 + 5698898 UCUUUCAGAUUAGUUCUCCAAA-----------UCAUGCUCAAAAACGAUAGAUCGAUCGAUCGAUCGUUCUUUCAGCCCACGUAUGUAUGUAUCUUAUGUAUGUAUCUUCGGAACUGAA .........((((((((.....-----------....(((.(((((((((.((((....)))).)))))).))).)))..(((((((((.......)))))))))......)))))))). ( -27.40, z-score = -2.58, R) >consensus UCUUUCAAAUUAGUUCUCCAAAA__________UCAUGCUCAAAAACGAUAGAUCGAUCGAUCGAUCGGUCUUUCAGCCCACGUAUGUAUGUAACUUAUAUAUGUAUCUUCGGAACUGAA ..................................(((((...........(((((((((....)))))))))..........)))))................................. (-10.60 = -11.85 + 1.25)

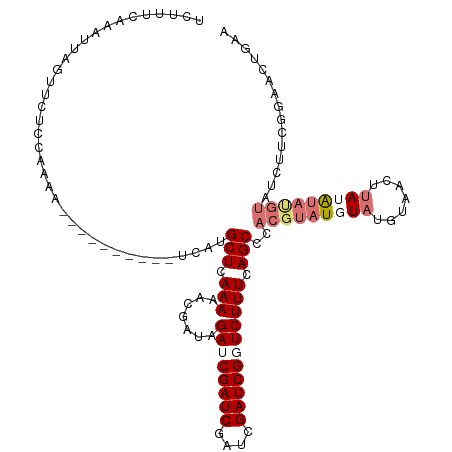
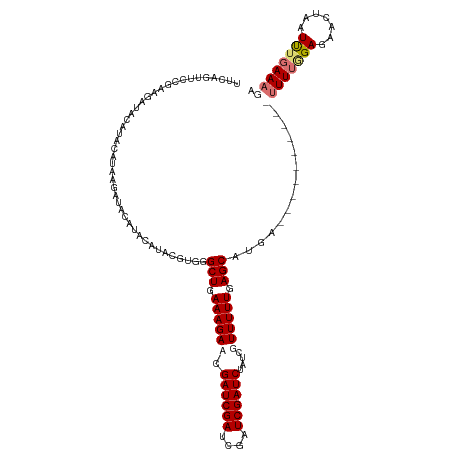
| Location | 10,431,459 – 10,431,578 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.42986 |
| G+C content | 0.37195 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -14.06 |
| Energy contribution | -15.50 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10431459 119 - 22422827 UUCAGUUCCGAAGAUACGUAUGUACAUAAGAUAUAUACGUGGGCUGAAAGACCGAUCGAUCGAUCGAUCUAUAGUUUUUGAGCAUGA-UUUCUCGUCUUUGGGAGAACUAAUUUGAAAGA (((((((((((((((((((((((((....).)))))))))..(((.((((((.((((((....))))))....)))))).)))....-......))))))))))........)))))... ( -35.30, z-score = -2.57, R) >droEre2.scaffold_4690 14048258 95 + 18748788 --------------------UUUAGUUUCAA-AGAUACGUGGGCUGAAAGACCGAUCGACCGAUCGAUC----GUUUUUGAGCAUGAUUUUCUCGUCUUUUAGAGAACUAAUUUGAAAGA --------------------.....((((((-(......((((((.((((((.((((((....))))))----)))))).)))......(((((........)))))))).))))))).. ( -26.90, z-score = -2.07, R) >droSec1.super_15 1131269 110 - 1954846 UUCAGUUCCGAAGAUACAUACAUAAGAUACAUACAUACGUGGGCUGAAAGAACGAUCGAUCGAUCGAUCUAUCGUUUUUGAGCAUGA----------UUUUUGAUGAUGAAUUUGAAAGA ((((....(((((((.(((..........(((......))).(((..(((((((((.((((....)))).))))))))).)))))))----------))))))....))))......... ( -26.00, z-score = -1.25, R) >droSim1.chrX_random 2949846 109 - 5698898 UUCAGUUCCGAAGAUACAUACAUAAGAUACAUACAUACGUGGGCUGAAAGAACGAUCGAUCGAUCGAUCUAUCGUUUUUGAGCAUGA-----------UUUGGAGAACUAAUCUGAAAGA ((((((((((((....(((..........(((......))).(((..(((((((((.((((....)))).))))))))).)))))).-----------))))))).......)))))... ( -26.91, z-score = -1.62, R) >consensus UUCAGUUCCGAAGAUACAUACAUAAGAUACAUACAUACGUGGGCUGAAAGAACGAUCGAUCGAUCGAUCUAUCGUUUUUGAGCAUGA__________UUUUGGAGAACUAAUUUGAAAGA ..........................................(((.((((((.((((((....))))))....)))))).))).............((((((((.......)))))))). (-14.06 = -15.50 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:32 2011