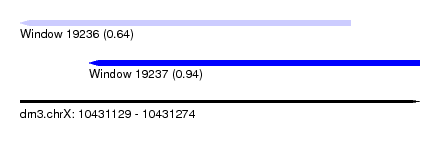
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,431,129 – 10,431,274 |
| Length | 145 |
| Max. P | 0.942729 |

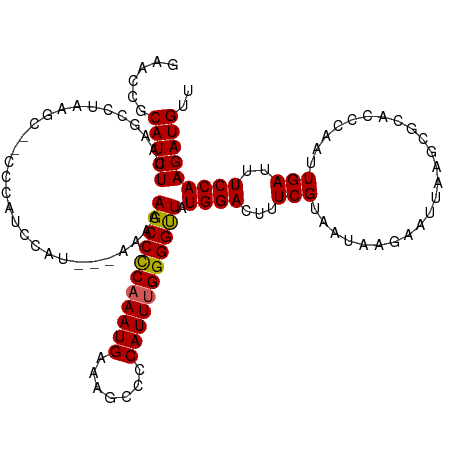
| Location | 10,431,129 – 10,431,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.85 |
| Shannon entropy | 0.19566 |
| G+C content | 0.38872 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10431129 120 - 22422827 CCAUCCAUAAAAAAAAGCCCCAAAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUUUACAACUCUAUGGAAACGAAUCGUU ...((((((.......((((((((((.......))))))))))))))))..(((((......((((((..........))))))(((((((.((.....))))).)))).)))))..... ( -29.01, z-score = -2.57, R) >droSim1.chrX_random 2949504 118 - 5698898 CCAUCCAU--AAAAAAGCCUCAAAUGAAAGCCCCAUUUGGGGCUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUUUACAACUCUAUGGAAACGAAUCGUU ...(((((--.....(((((((((((.......))))))))))))))))..(((((......((((((..........))))))(((((((.((.....))))).)))).)))))..... ( -28.30, z-score = -2.28, R) >droSec1.super_15 1130934 117 - 1954846 CCAUCCAU---AAAAAGCCUCAAAUGAAGGCCCCAUUUGGGGCUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUUUACAACUCUAUGGAAACGAAUCGUU ...(((((---....(((((((((((.......))))))))))))))))..(((((......((((((..........))))))(((((((.((.....))))).)))).)))))..... ( -28.70, z-score = -1.84, R) >droEre2.scaffold_4690 14047896 115 + 18748788 -CAGCUAU---AAAAAGCCCC-AAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAAGAAUUAAGCACACACAAUUGAUUUCCAAGAUGUUUACAACUCUAUGGGAACGAAUCGUU -..(((..---....((((((-((((.......)).)))))))).......(((.......)))...)))........(((.(((((((((.((.....))))).)))))))))...... ( -22.30, z-score = -0.32, R) >droYak2.chrX 18997419 101 - 21770863 -------------------CCAAAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAGGAAUAUAGCACACCCAAUUGAUUUCCAAGAUGUUUACAACUCUAUGGAAACGAAUCGUU -------------------...(((((.((((((....)))))).......(((((....((............)).......((((((((.((.....))))).))))))))))))))) ( -21.70, z-score = -0.70, R) >consensus CCAUCCAU___AAAAAGCCCCAAAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUUUACAACUCUAUGGAAACGAAUCGUU ...............(((((((((((.......)))))))))))...(((.(((((...............((......))..((((((((.((.....))))).))))))))))..))) (-21.70 = -22.14 + 0.44)

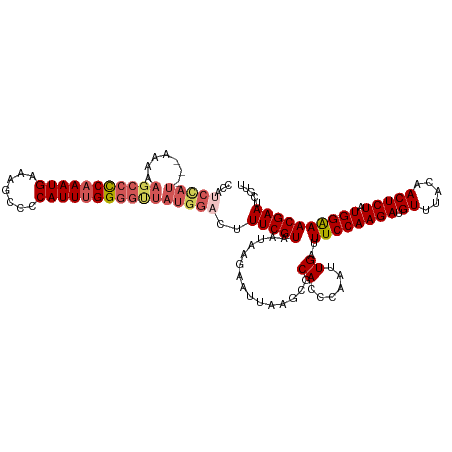
| Location | 10,431,154 – 10,431,274 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Shannon entropy | 0.26066 |
| G+C content | 0.41990 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.06 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10431154 120 - 22422827 GAAUCGCAUCUUAAGCCUAAGCUGCCCAUCCAUAAAAAAAAGCCCCAAAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUU .....(((((((.(((....)))((.(.((((((.......((((((((((.......))))))))))))))))..(((.......)))....).))...............))))))). ( -30.11, z-score = -2.78, R) >droSim1.chrX_random 2949529 117 - 5698898 GAACCGCAUCUCAAGCCUAAGCC-UCCAUCCAU--AAAAAAGCCUCAAAUGAAAGCCCCAUUUGGGGCUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUU .....(((((....((((.....-....(((((--.....(((((((((((.......))))))))))))))))..(((.......)))...)).))......(((.....)))))))). ( -26.30, z-score = -1.73, R) >droSec1.super_15 1130959 116 - 1954846 GAAACGCAUCUCAAGACUAAGCC-UCCAUCCAU---AAAAAGCCUCAAAUGAAGGCCCCAUUUGGGGCUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUU .....((((((............-....(((((---....(((((((((((.......))))))))))))))))............(((((((..........)))))))...)))))). ( -26.10, z-score = -1.25, R) >droEre2.scaffold_4690 14047921 113 + 18748788 GAACGCCAUCUUAAGCCUAAG---CCCAGCUAU---AAAAAGCCCC-AAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAAGAAUUAAGCACACACAAUUGAUUUCCAAGAUGUU ......((((((..((..(((---.((((((..---....)))...-......((((((....)))))).))).)))..)).....(((((((..........)))))))..)))))).. ( -23.30, z-score = -1.16, R) >droYak2.chrX 18997444 99 - 21770863 GAACCCCAUCUUGAGCCU---------------------AAGCCCCAAAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAGGAAUAUAGCACACCCAAUUGAUUUCCAAGAUGUU ......(((((((((((.---------------------.(((((((((((.......)))))))))))..)).))...((.(((....))).))................))))))).. ( -27.60, z-score = -2.80, R) >consensus GAACCGCAUCUUAAGCCUAAGC__CCCAUCCAU___AAAAAGCCCCAAAUGAAAGCCCCAUUUGGGGUUAUGGACUUUCGUAAUAAGAAUUAAGCGCACCCAAUUGAUUUCCAAGAUGUU ......(((((.............................(((((((((((.......))))))))))).((((...(((........................)))..))))))))).. (-22.34 = -22.06 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:29 2011