| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,413,028 – 10,413,153 |
| Length | 125 |
| Max. P | 0.990346 |

| Location | 10,413,028 – 10,413,153 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 136 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Shannon entropy | 0.50843 |
| G+C content | 0.48247 |
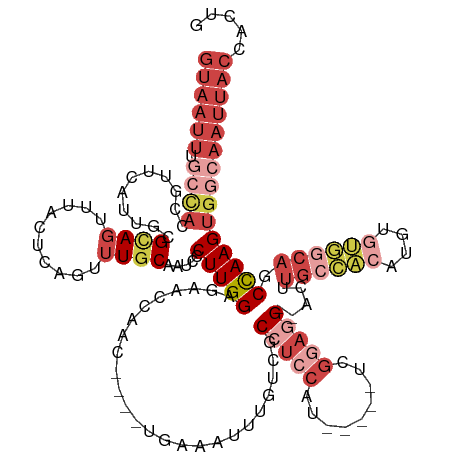
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
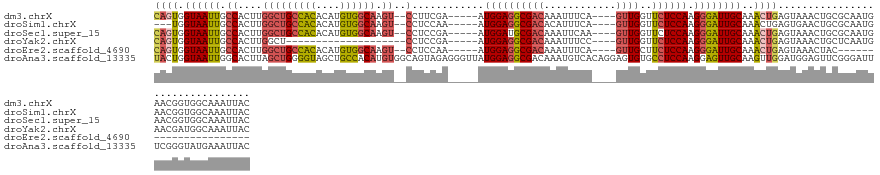
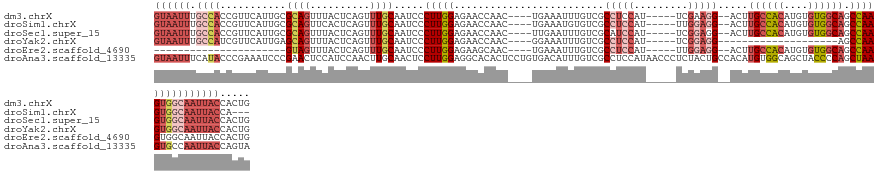
>dm3.chrX 10413028 125 + 22422827 CAGUGGUAAUUGCCACUUGGCUGCCACACAUGUGGCAAGU--CCUUCGA-----AUGGAGGCGACAAAUUUCA----GUUGGUUCUCCAAGGGAUUGCAAACUGAGUAAACUGCGCAAUGAACGGUGGCAAAUUAC .....((((((((((((.(((((((((....))))).)))--)((((..-----..))))(((......((((----(((.((..(((...)))..)).))))))).......))).......)))))).)))))) ( -42.52, z-score = -2.26, R) >droSim1.chrX 8091574 122 + 17042790 ---UGGUAAUUGCCACUUGGCUGCCACACAUGUGGCAAGU--CCUCCAA-----AUGGAGGCGACACAUUUCA----GUUGGUUCUCCAAGGGAUUGCAAACUGAGUGAACUGCGCAAUGAACGGUGGCAAAUUAC ---..((((((((((((.(((((((((....))))).)))--)((((..-----..))))(((......((((----(((.((..(((...)))..)).))))))).......))).......)))))).)))))) ( -46.12, z-score = -3.09, R) >droSec1.super_15 1120194 125 + 1954846 CAGUGGUAAUUGCCACUUGGCUGCCACACAUGUGGCAAGU--CCUCCGA-----AUGGAUGCGACAAAUUCAA----GUUGGUUCUCCAAGGGAUUGCAAACUGAGUAAACUGCGCAAUGAACGGUGGCAAAUUAC .....((((((((((((.(((((((((....))))).)))--).(((..-----..)))((((.((.(((((.----(((.((..(((...)))..)).))))))))....))))))......)))))).)))))) ( -39.70, z-score = -1.50, R) >droYak2.chrX 18987064 107 + 21770863 CAGUGGUAAUUGCCACUUGGCU--------------------CCUCCGA-----AUGGAGGCGACAAAUUUCC----GUUGGUUCUCCAAGGGAUUGCAAACUGAGUAAACUGCUCAAUGAACGAUGGCAAAUUAC .....((((((((((.(((((.--------------------(((((..-----..))))).........(((----.((((....)))).)))..))....(((((.....))))).....))))))).)))))) ( -33.10, z-score = -2.20, R) >droEre2.scaffold_4690 14036641 103 - 18748788 CAGUGGUAAUUGCCACUUGGCUGCCACACAUGUGGCAAGU--CCUCCAA-----AUGGAGGCGACAAAUUUCA----GUUGCUUCUCCAAGGGAUUGCAAACUGAGUAAACUAC---------------------- .((((((....))))))....((((((....)))))).(.--(((((..-----..)))))).......((((----(((((.(((.....)))..)).)))))))........---------------------- ( -36.40, z-score = -2.85, R) >droAna3.scaffold_13335 1047107 136 - 3335858 UACUGGUAAUUGGCACUUAGCUGGGGUAGCUGCCACAUGUGGCAGUAGAGGGUUAUGGAGGCGACAAAUGUCACAGGAGUGUGCCUCCAAGGAGUUGCAAGUUGGAUGGAGUUCGGGAUUUCGGGUAUGAAAUUAC ..((((...((..((..(((((...((((((((((....))))))).........((((((((((....)))(((....))))))))))......))).)))))..))))..))))(((((((....))))))).. ( -40.60, z-score = -0.91, R) >consensus CAGUGGUAAUUGCCACUUGGCUGCCACACAUGUGGCAAGU__CCUCCGA_____AUGGAGGCGACAAAUUUCA____GUUGGUUCUCCAAGGGAUUGCAAACUGAGUAAACUGCGCAAUGAACGGUGGCAAAUUAC ((((.(((((((((....)))((((((....))))))..................((((((((((............))))..))))))...))))))..))))................................ (-19.62 = -20.45 + 0.83)
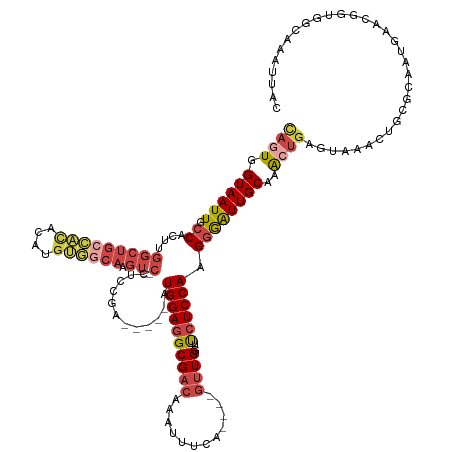
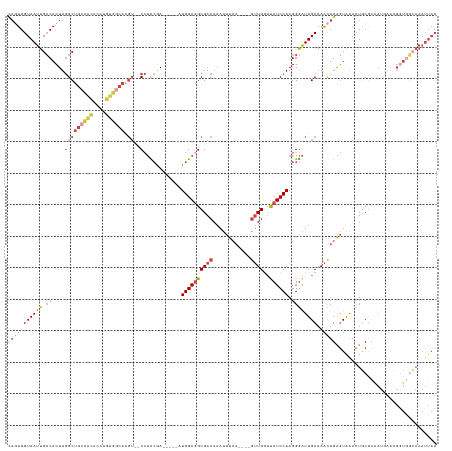
| Location | 10,413,028 – 10,413,153 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 136 |
| Reading direction | reverse |
| Mean pairwise identity | 73.11 |
| Shannon entropy | 0.50843 |
| G+C content | 0.48247 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -16.73 |
| Energy contribution | -19.97 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10413028 125 - 22422827 GUAAUUUGCCACCGUUCAUUGCGCAGUUUACUCAGUUUGCAAUCCCUUGGAGAACCAAC----UGAAAUUUGUCGCCUCCAU-----UCGAAGG--ACUUGCCACAUGUGUGGCAGCCAAGUGGCAAUUACCACUG ((((((.(((((((......((((((.....((((((.(...(((...)))...).)))----)))...))).)))......-----.))..((--.((.(((((....)))))))))..)))))))))))..... ( -37.62, z-score = -2.30, R) >droSim1.chrX 8091574 122 - 17042790 GUAAUUUGCCACCGUUCAUUGCGCAGUUCACUCAGUUUGCAAUCCCUUGGAGAACCAAC----UGAAAUGUGUCGCCUCCAU-----UUGGAGG--ACUUGCCACAUGUGUGGCAGCCAAGUGGCAAUUACCA--- ((((((.(((((........(((((......((((((.(...(((...)))...).)))----)))..)))))..(((((..-----..)))))--..(((((((....)))))))....)))))))))))..--- ( -41.80, z-score = -2.59, R) >droSec1.super_15 1120194 125 - 1954846 GUAAUUUGCCACCGUUCAUUGCGCAGUUUACUCAGUUUGCAAUCCCUUGGAGAACCAAC----UUGAAUUUGUCGCAUCCAU-----UCGGAGG--ACUUGCCACAUGUGUGGCAGCCAAGUGGCAAUUACCACUG ((((((.(((((.((((..(((((((.....((((.(((...(((...)))....))).----))))..))).))))(((..-----..)))))--))(((((((....)))))))....)))))))))))..... ( -39.10, z-score = -2.25, R) >droYak2.chrX 18987064 107 - 21770863 GUAAUUUGCCAUCGUUCAUUGAGCAGUUUACUCAGUUUGCAAUCCCUUGGAGAACCAAC----GGAAAUUUGUCGCCUCCAU-----UCGGAGG--------------------AGCCAAGUGGCAAUUACCACUG ((((((.(((...((..((((((.......))))))..))..(((.((((....)))).----))).(((((.(.(((((..-----..)))))--------------------.).))))))))))))))..... ( -34.50, z-score = -3.26, R) >droEre2.scaffold_4690 14036641 103 + 18748788 ----------------------GUAGUUUACUCAGUUUGCAAUCCCUUGGAGAAGCAAC----UGAAAUUUGUCGCCUCCAU-----UUGGAGG--ACUUGCCACAUGUGUGGCAGCCAAGUGGCAAUUACCACUG ----------------------((((((...((((((.((..(((...)))...)))))----))).........(((((..-----..)))))--...((((((....))))))(((....)))))))))..... ( -34.80, z-score = -2.44, R) >droAna3.scaffold_13335 1047107 136 + 3335858 GUAAUUUCAUACCCGAAAUCCCGAACUCCAUCCAACUUGCAACUCCUUGGAGGCACACUCCUGUGACAUUUGUCGCCUCCAUAACCCUCUACUGCCACAUGUGGCAGCUACCCCAGCUAAGUGCCAAUUACCAGUA (..(((((......)))))..)............((((((.......((((((((((....)))(((....))))))))))..........((((((....))))))........)).)))).............. ( -33.40, z-score = -3.44, R) >consensus GUAAUUUGCCACCGUUCAUUGCGCAGUUUACUCAGUUUGCAAUCCCUUGGAGAACCAAC____UGAAAUUUGUCGCCUCCAU_____UCGGAGG__ACUUGCCACAUGUGUGGCAGCCAAGUGGCAAUUACCACUG ((((((.((((...........((((..........)))).....(((((.........................(((((.........))))).....((((((....)))))).)))))))))))))))..... (-16.73 = -19.97 + 3.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:27 2011