
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,400,076 – 10,400,176 |
| Length | 100 |
| Max. P | 0.630396 |

| Location | 10,400,076 – 10,400,176 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 52.40 |
| Shannon entropy | 0.65058 |
| G+C content | 0.29637 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -5.53 |
| Energy contribution | -4.65 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
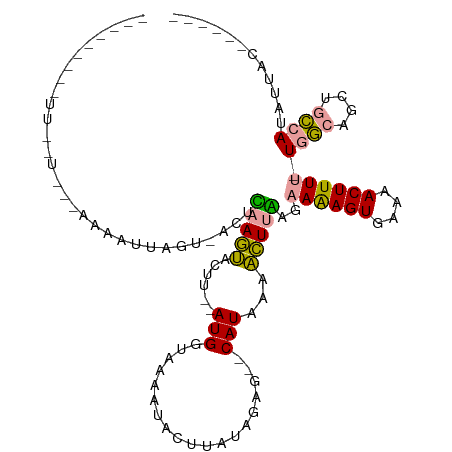
>dm3.chrX 10400076 100 - 22422827 ------AAAUUUGUAUGAAUAUUAGCAACUCCAGUACUU--AUGGUAAAUUAUUUCUGGAG--CAUAAAACUUGCGAAAAGUGAAAACUUUU-UCGCAGCUGCCAUAUUAC------ ------...........(((((((((..((((((.....--((.....)).....))))))--.........((((((((((....)).)))-)))))))))..)))))..------ ( -24.30, z-score = -2.94, R) >droAna3.scaffold_13335 1033800 117 + 3335858 ACUUUAUUUUUAAUGCAAACAUCAGUUACUGAAAGCCUCCAAUGAUGAUAUGCCCAUAAAAUCCAUCAACUUUCAGCAGAGUGAAAACUUUUAUGACAGCUUUCAUUUUUCAAAAAC .............................(((((((......(((((................)))))....(((..(((((....)))))..)))..)))))))............ ( -16.89, z-score = -0.80, R) >droYak2.chrX 18974016 87 - 21770863 ------------------AAAUUUAU-AUUUAAGUAUUU--AUGGGAAAACACUUUUAGAG--CAUUAAACUUAUGAAAAGUGAAAACUUUU-UGGCAUCUGCCAAAUUAC------ ------------------...(((((-((((((...(((--(..((......))..)))).--..)))))..))))))((((....))))((-((((....))))))....------ ( -15.00, z-score = -1.63, R) >consensus _________UU__U___AAAAUUAGU_ACUCAAGUACUU__AUGGUAAAAUACUUAUAGAG__CAUAAAACUUAAGAAAAGUGAAAACUUUU_UGGCAGCUGCCAUAUUAC______ ..............................(((((..................................)))))..((((((....)))))).((((....))))............ ( -5.53 = -4.65 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:25 2011