
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,388,053 – 10,388,163 |
| Length | 110 |
| Max. P | 0.752339 |

| Location | 10,388,053 – 10,388,163 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 49.29 |
| Shannon entropy | 0.88169 |
| G+C content | 0.54430 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -10.03 |
| Energy contribution | -10.32 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10388053 110 - 22422827 UUGCGAGCGAUUCCUAC----------AGGUAGUGGCUUGGAUGGACCAAUGUCCAAUUACAUUACGCUGGGAUUGGG--CUACCUGUGG---------GUAGUGGGUGGAAUACCAAUCGAAGCCAGCGG ...(((((.(((((...----------.)).))).)))))..(((((....))))).........((((((((((((.--(((((..(..---------...)..)))))....))))))....)))))). ( -43.60, z-score = -2.85, R) >droSec1.super_15 1095440 105 - 1954846 UUGCGAGCGAUUCAUAC----------AGGUAGUGGCUUGGAUGGACCAAUGUCCAA-AACAUUACGCUGGGAUUGGG--CUACCUGGG------------AGUGAGUGGAAUACCAAUCCAAGCCAGCG- ..((..((........(----------((((((((..(((((((......)))))))-..)))))).)))(((((((.--(((((....------------...).))))....)))))))..))..)).- ( -38.60, z-score = -2.67, R) >droYak2.chrX 18961397 127 - 21770863 UUGCCAGCGAUUCCUACCAGGUAGUAGUAGUAGUGGCUUCGAUGGACCAAUGUCCAA-GACACUAAACGGGGAUUGGG--CUACCUGGGCGGACUACCUGGUGGGUGUGCAAAGCCAAUCCAGGCCAGCG- ..((..(((...(((((((((((((.((....(((((.((....))(((((.(((..-..........))).))))))--))))....))..)))))))))))))..)))...(((......)))..)).- ( -50.50, z-score = -1.72, R) >droEre2.scaffold_4690 14008523 106 + 18748788 UUGCCAGCGAUUCCUAC----------AGGUAGUGGCUGGGAUGGGCCAAUGUCCAA-AAAAUCAAACUGGGAUUGGA--CUACCUGAAG-----------UGGGUGUGCAAAGGCAAUCCCGGCCAGCG- ......((.....(((.----------...)))((((((((((..(((...((((((-....((.....))..)))))--)(((((....-----------.)))))......))).)))))))))))).- ( -38.90, z-score = -1.40, R) >droAna3.scaffold_13335 1020778 84 + 3335858 ----------------------------------GGUUUCUACACUUGUUGGUUCGAAAAGAACCUCCGAUUGAUGAA--UCAUCCUGG----------CUAGGGGGUGGUACCUGGACUUUGGGCAGGU- ----------------------------------(((((...((.(((..(((((.....)))))..))).))..)))--))..((((.----------((((((..(((...)))..)))))).)))).- ( -25.50, z-score = -0.43, R) >dp4.chrXL_group1e 5733900 102 - 12523060 -------------------UUUCGGGCGACUUGGGACUGGGAUUCACCGCGGUCGA--UGGCUCAUACUGGGGCUGGGGCCUGCCCCCGA-------UUUCCACUUGAGUCAUGGCAUCCUCGGCGACGA- -------------------..(((..((.((.((((..((......))...(((.(--(((((((...((((.(.((((....)))).).-------..))))..))))))))))).)))).)))).)))- ( -37.70, z-score = 0.23, R) >consensus UUGC_AGCGAUUCCUAC__________AGGUAGUGGCUUGGAUGGACCAAUGUCCAA_AACAUCAAACUGGGAUUGGG__CUACCUGGG____________AGGGUGUGGAAUGCCAAUCCAGGCCAGCG_ ..................................(((((...(((((....)))))..............(((((((....(((......................))).....))))))))))))..... (-10.03 = -10.32 + 0.28)

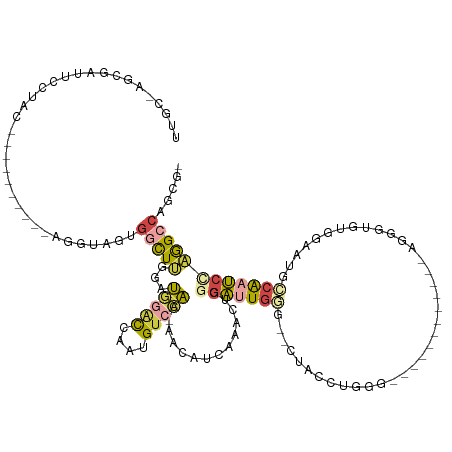
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:23 2011