
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,337,843 – 10,337,937 |
| Length | 94 |
| Max. P | 0.834242 |

| Location | 10,337,843 – 10,337,937 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 60.50 |
| Shannon entropy | 0.64638 |
| G+C content | 0.50733 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.04 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
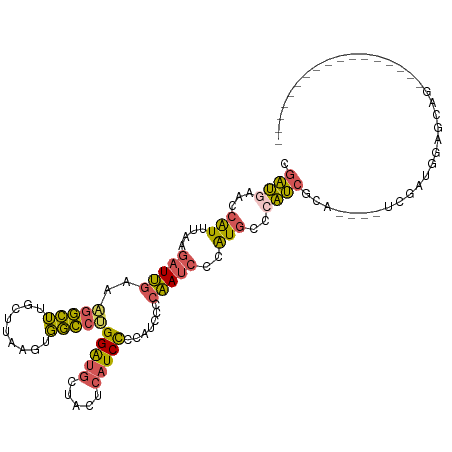
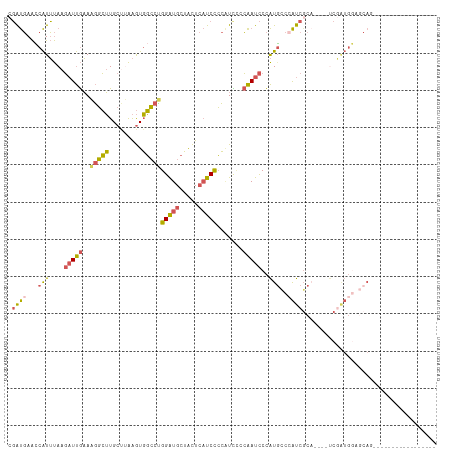
>dm3.chrX 10337843 94 + 22422827 CGAUGAACCAUUUAAGAUUGAAAGGCUUGCUAAAGUGGCCCGGAUGCUACUCAUCCCCCUUUCCAAUCCCAUGCCCAUCGCA----UCGCUGGAGCAG----------------- ...............((((((((((...(((.....)))..(((((.....))))).))))).)))))...((((((.((..----.)).))).))).----------------- ( -22.10, z-score = -0.38, R) >droYak2.chrX 18911336 114 + 21770863 CGAUGAAGCAUUUAAGAUUGAAAGGCUUGCCUAAGUGGCCUGGAUGCUACUCAUCCCCAUC-CCAAUCCCAUUCCAAUCCCCAUACCCAAUGCUUCAAAACCAUGCCCAUCGCAU ...(((((((((...(((((..(((((.........)))))(((((.....))))).....-............))))).........))))))))).....((((.....)))) ( -24.60, z-score = -1.75, R) >droSec1.super_15 1045363 94 + 1954846 CGAUGAACCAUUUAAGAUUGAAAGGCUUGCUUAAGUGGCCUGGAUGCUACUCAUCCCCAUCCCCAAUCCCAUGCCCAUCGCA----UCGAUGGAGCAG----------------- .((((..(((((((((...(.....)...)))))))))...(((((.....))))).))))..........(((((((((..----.)))))).))).----------------- ( -28.30, z-score = -2.37, R) >droSim1.chrX 8042527 94 + 17042790 CGAUGAACCAUUUAAGAUUGAAAGGCUUGCUUAAGUGGCCUGGAUGCUACUCAUCCCCAUCCCCAAUCCCAUGCCCAUCGCA----UCGAUGGAGCAG----------------- .((((..(((((((((...(.....)...)))))))))...(((((.....))))).))))..........(((((((((..----.)))))).))).----------------- ( -28.30, z-score = -2.37, R) >droVir3.scaffold_12932 191974 86 + 2102469 -UGCCAGUCGUGCAAUUUCACAUUCAGUGCCAGGAUUUGUGGGGUGUAGUU--UCUGAGUUUGGGAUUCUGUGUUUGCCGCU-CGCCCAG------------------------- -.((.((.((.((((...((((...(((.(((((.....(((((......)--))))..))))).))).)))).))))))))-.))....------------------------- ( -19.50, z-score = 1.26, R) >consensus CGAUGAACCAUUUAAGAUUGAAAGGCUUGCUUAAGUGGCCUGGAUGCUACUCAUCCCCAUCCCCAAUCCCAUGCCCAUCGCA____UCGAUGGAGCAG_________________ .((((...(((....(((((..(((((.........)))))(((((.....))))).......)))))..)))..)))).................................... (-11.64 = -11.04 + -0.60)

| Location | 10,337,843 – 10,337,937 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 60.50 |
| Shannon entropy | 0.64638 |
| G+C content | 0.50733 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.776378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10337843 94 - 22422827 -----------------CUGCUCCAGCGA----UGCGAUGGGCAUGGGAUUGGAAAGGGGGAUGAGUAGCAUCCGGGCCACUUUAGCAAGCCUUUCAAUCUUAAAUGGUUCAUCG -----------------.(((....))).----..((((((((.((((((((.(((((.(((((.....)))))..((.......))...)))))))))))))....)))))))) ( -30.40, z-score = -0.80, R) >droYak2.chrX 18911336 114 - 21770863 AUGCGAUGGGCAUGGUUUUGAAGCAUUGGGUAUGGGGAUUGGAAUGGGAUUGG-GAUGGGGAUGAGUAGCAUCCAGGCCACUUAGGCAAGCCUUUCAAUCUUAAAUGCUUCAUCG ((((.....)))).....(((((((((.......((((((((((.((......-.....(((((.....)))))..(((.....)))...)))))))))))).)))))))))... ( -35.50, z-score = -1.15, R) >droSec1.super_15 1045363 94 - 1954846 -----------------CUGCUCCAUCGA----UGCGAUGGGCAUGGGAUUGGGGAUGGGGAUGAGUAGCAUCCAGGCCACUUAAGCAAGCCUUUCAAUCUUAAAUGGUUCAUCG -----------------.(((.((((((.----..)))))))))((((((((..(....(((((.....))))).(((...........))))..))))))))............ ( -29.60, z-score = -0.57, R) >droSim1.chrX 8042527 94 - 17042790 -----------------CUGCUCCAUCGA----UGCGAUGGGCAUGGGAUUGGGGAUGGGGAUGAGUAGCAUCCAGGCCACUUAAGCAAGCCUUUCAAUCUUAAAUGGUUCAUCG -----------------.(((.((((((.----..)))))))))((((((((..(....(((((.....))))).(((...........))))..))))))))............ ( -29.60, z-score = -0.57, R) >droVir3.scaffold_12932 191974 86 - 2102469 -------------------------CUGGGCG-AGCGGCAAACACAGAAUCCCAAACUCAGA--AACUACACCCCACAAAUCCUGGCACUGAAUGUGAAAUUGCACGACUGGCA- -------------------------....((.-((((((((.....................--.........(((.......)))(((.....)))...)))).)).)).)).- ( -14.50, z-score = 0.97, R) >consensus _________________CUGCUCCAUCGA____UGCGAUGGGCAUGGGAUUGGAGAUGGGGAUGAGUAGCAUCCAGGCCACUUAAGCAAGCCUUUCAAUCUUAAAUGGUUCAUCG ....................................(((((((.((((((((((.....(((((.....)))))((((...........))))))))))))))....))))))). (-17.36 = -17.72 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:16 2011