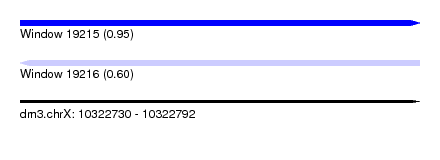
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,322,730 – 10,322,792 |
| Length | 62 |
| Max. P | 0.949090 |

| Location | 10,322,730 – 10,322,792 |
|---|---|
| Length | 62 |
| Sequences | 5 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 55.21 |
| Shannon entropy | 0.75426 |
| G+C content | 0.56215 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -6.76 |
| Energy contribution | -7.24 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
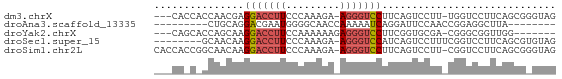
>dm3.chrX 10322730 62 + 22422827 ---CACCACCAACGAGGACCUUCCCAAAGA-AGGGUCCUUCAGUCCUU-UGGUCCUUCAGCGGGUAG ---.(((..(...(((((((........((-((....)))).......-.)))))))..)..))).. ( -18.59, z-score = -0.87, R) >droAna3.scaffold_13335 935423 50 - 3335858 ---------CUGCAGGACGAAUGGGGCAACCAAAAAUCAGGAUUCCAACCGGAGGCUUA-------- ---------..((..........((....))...........((((....))))))...-------- ( -8.80, z-score = 0.67, R) >droYak2.chrX 18895491 56 + 21770863 ---CAGCACCAGCAAGGACCUUCCAAAAAAGAGGGUCCUUCGGUGCGA-CGGGCGGUUGG------- ---..(((((.(.((((((((((.......))))))))))))))))..-...........------- ( -24.50, z-score = -2.92, R) >droSec1.super_15 1017092 58 + 1954846 --------GCAACAAGGACCUUCCCAAAGA-AGGGUCCAUCAGUCCUUUCGGUCCUUCAGCGUGUAG --------((...(((((((........((-((((.(.....).)))))))))))))..))...... ( -17.00, z-score = -2.60, R) >droSim1.chr2L 20296088 65 + 22036055 CACCACCGGCAACAAGGACCUUCCCAAAGA-AGGGUCCUUCAGUCCUU-CGGUCCUUCAGCGGGUAG ....(((.((...(((((((........((-((((........)))))-))))))))..)).))).. ( -22.30, z-score = -2.28, R) >consensus ___CA_C_CCAACAAGGACCUUCCCAAAGA_AGGGUCCUUCAGUCCUU_CGGUCCUUCAGCG_GUAG ...............(((((((.........)))))))............................. ( -6.76 = -7.24 + 0.48)

| Location | 10,322,730 – 10,322,792 |
|---|---|
| Length | 62 |
| Sequences | 5 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 55.21 |
| Shannon entropy | 0.75426 |
| G+C content | 0.56215 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -6.16 |
| Energy contribution | -5.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
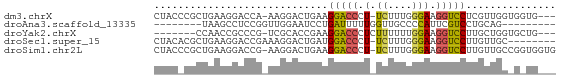
>dm3.chrX 10322730 62 - 22422827 CUACCCGCUGAAGGACCA-AAGGACUGAAGGACCCU-UCUUUGGGAAGGUCCUCGUUGGUGGUG--- ....((((..(((((((.-.......((((....))-))........))))))..)..))))..--- ( -21.49, z-score = -0.77, R) >droAna3.scaffold_13335 935423 50 + 3335858 --------UAAGCCUCCGGUUGGAAUCCUGAUUUUUGGUUGCCCCAUUCGUCCUGCAG--------- --------...((...((..(((...((........)).....)))..))....))..--------- ( -7.40, z-score = 0.89, R) >droYak2.chrX 18895491 56 - 21770863 -------CCAACCGCCCG-UCGCACCGAAGGACCCUCUUUUUUGGAAGGUCCUUGCUGGUGCUG--- -------...........-..(((((((((((((.(((.....))).)))))))..))))))..--- ( -22.90, z-score = -3.31, R) >droSec1.super_15 1017092 58 - 1954846 CUACACGCUGAAGGACCGAAAGGACUGAUGGACCCU-UCUUUGGGAAGGUCCUUGUUGC-------- ......((..(((((((....))..........(((-((.....))))))))))...))-------- ( -16.00, z-score = -0.85, R) >droSim1.chr2L 20296088 65 - 22036055 CUACCCGCUGAAGGACCG-AAGGACUGAAGGACCCU-UCUUUGGGAAGGUCCUUGUUGCCGGUGGUG (((((.((..(((((((.-.......((((....))-))........)))))))...)).))))).. ( -25.59, z-score = -1.98, R) >consensus CUAC_CGCUGAAGGACCG_AAGGACUGAAGGACCCU_UCUUUGGGAAGGUCCUUGUUGC_G_UG___ ...............((....))......(((((...((....))..)))))............... ( -6.16 = -5.88 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:11 2011