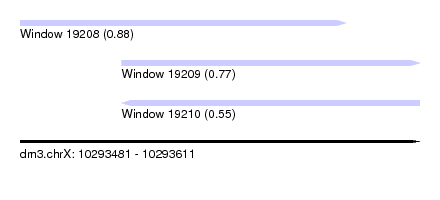
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,293,481 – 10,293,611 |
| Length | 130 |
| Max. P | 0.876459 |

| Location | 10,293,481 – 10,293,587 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Shannon entropy | 0.23375 |
| G+C content | 0.34147 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -16.13 |
| Energy contribution | -17.80 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10293481 106 + 22422827 UGGUCAUGAAAAUUCAAAUUCCUAUACAACUAU----ACAUAUUAUUGAAAAUAUGUUGAAAUUAGCCUUUUGCCAGCAAAUGGCUCUGCACAUGGCCAAAAUGCGAUGA ((((((((....(((((.....((((....)))----)((((((......)))))))))))...((((.((((....)))).)))).....))))))))........... ( -24.10, z-score = -1.77, R) >droSim1.chrX 8017087 92 + 17042790 UGGUCCUAAAAAUCCAAAUUCCUAUAGAACUAGGAAAACAUAUUAUUGAAAAUAUGU------------------AGCAAAUGGCUGUGCACAUGGCCAAAAUGCGAUGA (((..........)))..((((((......)))))).(((((((......)))))))------------------.(((..(((((((....)))))))...)))..... ( -21.80, z-score = -2.43, R) >droSec1.super_15 989761 92 + 1954846 UGGUCAUAAAAAUCCAAAUUCCUAUAGAACUAGGAAAACAUAUUAUUGAAAAUAUGU------------------AGCAAAUGGCUGUGCACAUGGCCAAAAUGCGAUGA ..(((.............((((((......)))))).(((((((......)))))))------------------.(((..(((((((....)))))))...)))))).. ( -22.50, z-score = -2.68, R) >consensus UGGUCAUAAAAAUCCAAAUUCCUAUAGAACUAGGAAAACAUAUUAUUGAAAAUAUGU__________________AGCAAAUGGCUGUGCACAUGGCCAAAAUGCGAUGA ..(((.............((((((......)))))).(((((((......)))))))...................(((..(((((((....)))))))...)))))).. (-16.13 = -17.80 + 1.67)

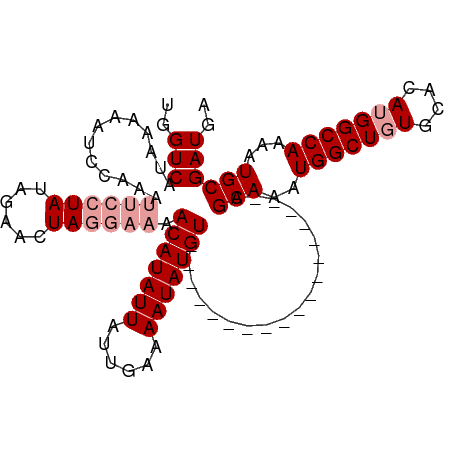
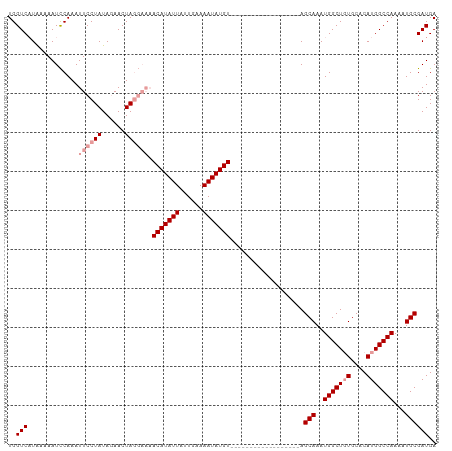
| Location | 10,293,514 – 10,293,611 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Shannon entropy | 0.22039 |
| G+C content | 0.34855 |
| Mean single sequence MFE | -18.87 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10293514 97 + 22422827 ---ACAUAUUAUUGAAAAUAUGUUGAAAUUAGCCUUUUGCCAGCAAAUGGCUCUGCACAUGGCCAAAAUGCGAUGACAGAAGAUGUUGUAUGUGAAAAAU ---(((((((......))))))).......((((.((((....)))).))))...(((((.((......)).(..(((.....)))..))))))...... ( -19.60, z-score = -0.44, R) >droSim1.chrX 8017121 82 + 17042790 AAAACAUAUUAUUGAAAAUAUGU------------------AGCAAAUGGCUGUGCACAUGGCCAAAAUGCGAUGACAGAAGAUGUUGUGUGUGGAAAAU ...(((((((......)))))))------------------.(((..(((((((....)))))))...))).(..(((.....)))..)........... ( -18.50, z-score = -1.58, R) >droSec1.super_15 989795 82 + 1954846 AAAACAUAUUAUUGAAAAUAUGU------------------AGCAAAUGGCUGUGCACAUGGCCAAAAUGCGAUGACAGAAGAUGUUGUAUGUGGAAAAU ...(((((((......)))))))------------------.(((..(((((((....)))))))...))).(..(((.....)))..)........... ( -18.50, z-score = -1.87, R) >consensus AAAACAUAUUAUUGAAAAUAUGU__________________AGCAAAUGGCUGUGCACAUGGCCAAAAUGCGAUGACAGAAGAUGUUGUAUGUGGAAAAU ...(((((((......)))))))...................(((..(((((((....)))))))...))).(..(((.....)))..)........... (-17.03 = -17.37 + 0.33)



| Location | 10,293,514 – 10,293,611 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Shannon entropy | 0.22039 |
| G+C content | 0.34855 |
| Mean single sequence MFE | -15.13 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10293514 97 - 22422827 AUUUUUCACAUACAACAUCUUCUGUCAUCGCAUUUUGGCCAUGUGCAGAGCCAUUUGCUGGCAAAAGGCUAAUUUCAACAUAUUUUCAAUAAUAUGU--- ....................((((((((.((......)).))).)))))(((.((((....)))).)))........(((((((......)))))))--- ( -20.40, z-score = -1.65, R) >droSim1.chrX 8017121 82 - 17042790 AUUUUCCACACACAACAUCUUCUGUCAUCGCAUUUUGGCCAUGUGCACAGCCAUUUGCU------------------ACAUAUUUUCAAUAAUAUGUUUU .............................(((...((((..((....))))))..))).------------------(((((((......)))))))... ( -12.50, z-score = -1.54, R) >droSec1.super_15 989795 82 - 1954846 AUUUUCCACAUACAACAUCUUCUGUCAUCGCAUUUUGGCCAUGUGCACAGCCAUUUGCU------------------ACAUAUUUUCAAUAAUAUGUUUU .............................(((...((((..((....))))))..))).------------------(((((((......)))))))... ( -12.50, z-score = -1.53, R) >consensus AUUUUCCACAUACAACAUCUUCUGUCAUCGCAUUUUGGCCAUGUGCACAGCCAUUUGCU__________________ACAUAUUUUCAAUAAUAUGUUUU .............................(((...((((..........))))..)))...................(((((((......)))))))... (-11.77 = -11.77 + 0.00)

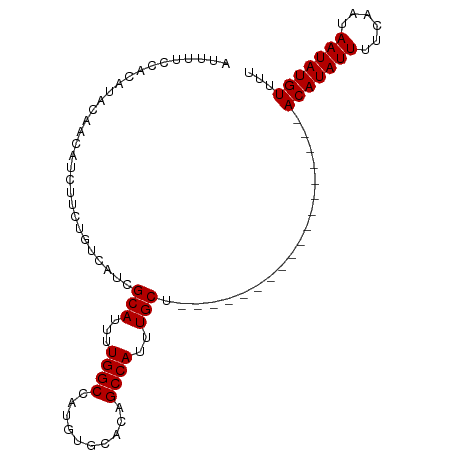
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:31:06 2011