| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,816,678 – 10,816,773 |
| Length | 95 |
| Max. P | 0.774784 |

| Location | 10,816,678 – 10,816,773 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Shannon entropy | 0.48611 |
| G+C content | 0.44187 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -11.99 |
| Energy contribution | -11.43 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
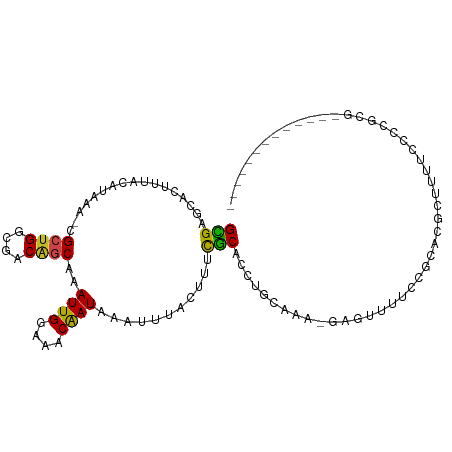
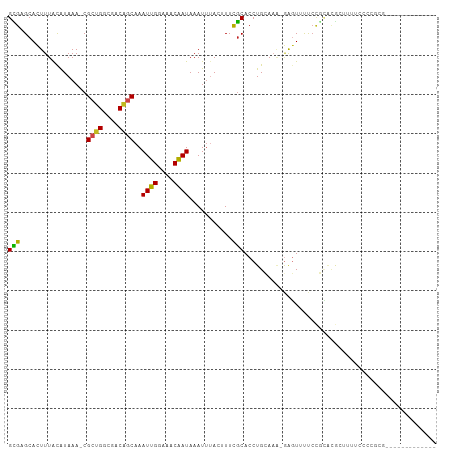
>dm3.chr2L 10816678 95 + 23011544 GCGAGCACUUUACAUAAA-CGCUGGCGACAGCAAAUUGGAAGCGAUAAAUUUACUUUCGCACCUGCAAAAGAGUUUUCCGCGCGAUUUUCCCCGCG------------- (((((.(((((.......-.((((....)))).....(((((..(.....)..)))))((....))...))))).)).)))(((........))).------------- ( -23.80, z-score = -0.91, R) >droGri2.scaffold_15126 2047740 87 + 8399593 GUGUGCACUUUAUGUAAACCGAUGACAGCGGCAAAUUGAAAACAAUAAAUUAA-AUUCGCACCUGAAAACGAGUGCCAGUGCAACACA--------------------- (((((((((....(((...((.(..(((..((.((((...............)-))).))..)))..).))..))).))))).)))).--------------------- ( -18.56, z-score = -0.96, R) >droMoj3.scaffold_6500 13550256 86 - 32352404 GCGUGCAGUUUACAUAAAACGACGGCGACAGCAAAUUGAAAACAAUAAAUAAA-AUUCGCACCUGAAAGGUGUUUCCCCU-AUGCCCA--------------------- ((.((..((((.....))))..))))....(((.((((....)))).......-....(((((.....))))).......-.)))...--------------------- ( -12.80, z-score = 0.57, R) >droVir3.scaffold_12963 4401463 96 - 20206255 GUGUGCACUUUACAUAAAACGCUGGCGACAGCAAAUUGAAAACAAUAAAUUUA-AUUCGCACCUGAAAAGAGUUUCCCCAGACGCCCUACACACACA------------ (((((...............((((....)))).....................-....((..((....)).(((......)))))....)))))...------------ ( -17.80, z-score = -1.79, R) >droWil1.scaffold_180703 376266 93 - 3946847 GUGUGCACUUUGCAUAAA-CGCUGGCGACAGCAAAUUGAAAACGAUAAAUUUA-UUUCACACCUGAAAAAGAGUUUCCUCUCGAUUGGUCCCCCU-------------- .(((((.....)))))..-.((((....)))).((((((..............-(((((....)))))..(((....))))))))).........-------------- ( -18.70, z-score = -1.12, R) >dp4.chr4_group4 3021561 92 + 6586962 GCGAGCACUUUGCAUAAA-CGCUGGCGACAGCAAAUUGGAAACAAUAAAUUUAUUUUUGCACCUGCA---GAGUUUUCCGCACACUUUUCCGCUUU------------- (((((.((((((((....-.((((....))))..((((....)))).................))))---)))).)).)))...............------------- ( -26.70, z-score = -3.21, R) >droPer1.super_10 2036248 92 + 3432795 GCGAGCACUUUGCAUAAA-CGCUGGCGACAGCAAAUUGGAAACAAUAAAUUUAUUUUUGCACCUGCA---GAGUUUUCCGCACACUUUUCCGCUUU------------- (((((.((((((((....-.((((....))))..((((....)))).................))))---)))).)).)))...............------------- ( -26.70, z-score = -3.21, R) >droSim1.chr2L 10623442 95 + 22036055 GUGAGCACUUUACAUAAA-CGCUGGCGACAGCAAAUUGGAAGCGAUAAAUUUACUUUCGCACCUGCAAAAGAGUUUUCCGCGCGAUUUUCCCCGCG------------- ....((............-.((((....)))).....((((((((...........))))((((.....)).)).))))))(((........))).------------- ( -22.60, z-score = -0.69, R) >droSec1.super_3 6234934 95 + 7220098 GCGAGCACUUUACAUAAA-CGCUGGCGACAGCAAAUUGGAAGCGAUAAAUUUACUUUCGCACCUGCAAAAGAGUUUUCCGUGCGAUUUUCCCCGCG------------- (((.((((..........-.((((....)))).....((((((((...........))))((((.....)).)).))))))))(.....)..))).------------- ( -24.60, z-score = -1.37, R) >droYak2.chr2L 7223763 94 + 22324452 GCGAGCACUUUACAUAAA-CGCUGGCGACAGCAAAUUGGAAACGAUAAAUUUACUUUCGCACCUGCAAA-GAGUUUUCCGCACGAUUUUCCCCGCG------------- (((((.(((((.......-.((((....))))..((((....))))............((....))..)-)))).)).)))...............------------- ( -20.30, z-score = -1.02, R) >droEre2.scaffold_4929 12027323 94 - 26641161 GCGAGCACUUUACAUAAA-CGCUGGCGACAGCAAAUUGGAAACAAUAAAUUUACUUUCGCACCUGCAAA-GAGUUUUCCGCGCGAUUUUCCCCGCG------------- (((((.(((((.......-.((((....))))..((((....))))............((....))..)-)))).)).)))(((........))).------------- ( -24.30, z-score = -2.00, R) >droAna3.scaffold_12916 14669312 107 + 16180835 GUGUGCACUUUACAUAAA-CGCUGGCGACAGCAAAUUGGAAACAAUAAAUUUACUUUUGCACCUGCAAA-GAGUUUUCCGCACGCUUUUCCCUUAAACCAGGUCGCGAA ((((((............-.((((....))))..((((....))))......((((((((....)).))-)))).....)))))).....(((......)))....... ( -27.90, z-score = -2.02, R) >consensus GCGAGCACUUUACAUAAA_CGCUGGCGACAGCAAAUUGGAAACAAUAAAUUUACUUUCGCACCUGCAAA_GAGUUUUCCGCACGCUUUUCCCCGCG_____________ (((.................((((....))))..((((....))))...........)))................................................. (-11.99 = -11.43 + -0.57)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:06 2011