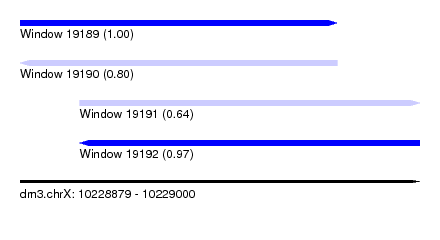
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,228,879 – 10,229,000 |
| Length | 121 |
| Max. P | 0.996178 |

| Location | 10,228,879 – 10,228,975 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Shannon entropy | 0.17275 |
| G+C content | 0.46665 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -21.77 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
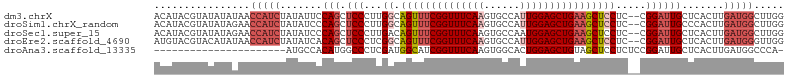
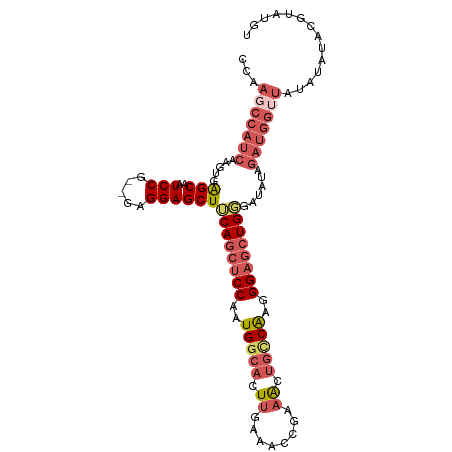
>dm3.chrX 10228879 96 + 22422827 -----CCCCCGCCACUACCAUCUACAUACGUAUAUAUAACCAUCUAUAUUCCAGCUCCCUUGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCU -----.....((.....................(((((......)))))..(((((((..(((((.((........)).)))))..)))))))..)).... ( -23.50, z-score = -2.90, R) >droSim1.chrX_random 589702 101 + 5698898 CCCCACCACCAUCUACAUACUAUACAUACGUAUAUAGAACCAUCUAUAUCCCAGCUCCCUUGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCU ...........((((.((((.........)))).)))).............(((((((..(((((.((........)).)))))..)))))))........ ( -26.80, z-score = -3.84, R) >droSec1.super_15 931810 101 + 1954846 CCCCACCACCAUCUACAUACUAUACAUACGUAUAUAGAACCAUCUAUAUCCCAGCUCCCUUGACAGUUUCGGUUUCAAGUGCCAAUGGAGCUGAAGCUCCU ...........((((.((((.........)))).)))).............(((((((.(((.((.((........)).)).))).)))))))........ ( -20.50, z-score = -2.31, R) >consensus CCCCACCACCAUCUACAUACUAUACAUACGUAUAUAGAACCAUCUAUAUCCCAGCUCCCUUGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCU .............................(.(((((((....))))))).)(((((((..(((((.((........)).)))))..)))))))........ (-21.77 = -22.43 + 0.67)

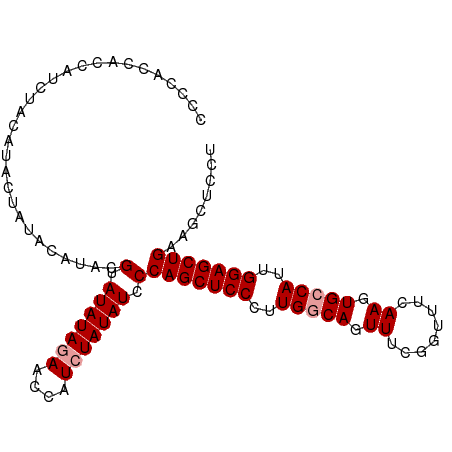
| Location | 10,228,879 – 10,228,975 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Shannon entropy | 0.17275 |
| G+C content | 0.46665 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -25.93 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10228879 96 - 22422827 AGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCAAGGGAGCUGGAAUAUAGAUGGUUAUAUAUACGUAUGUAGAUGGUAGUGGCGGGGG----- .(((((....)))))..............(((..(((((((((....)).............(((((((....))))))).)))))))..)))...----- ( -28.30, z-score = -1.56, R) >droSim1.chrX_random 589702 101 - 5698898 AGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCAAGGGAGCUGGGAUAUAGAUGGUUCUAUAUACGUAUGUAUAGUAUGUAGAUGGUGGUGGGG ......(((((((((..(((((.((........)).)))))..))))))))).((((((....))))))(((((((.....)))))))............. ( -31.60, z-score = -1.88, R) >droSec1.super_15 931810 101 - 1954846 AGGAGCUUCAGCUCCAUUGGCACUUGAAACCGAAACUGUCAAGGGAGCUGGGAUAUAGAUGGUUCUAUAUACGUAUGUAUAGUAUGUAGAUGGUGGUGGGG ......(((((((((.((((((.((........)).)))))).))))))))).((((((....))))))(((((((.....)))))))............. ( -29.70, z-score = -1.52, R) >consensus AGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCAAGGGAGCUGGGAUAUAGAUGGUUCUAUAUACGUAUGUAUAGUAUGUAGAUGGUGGUGGGG ......(((((((((..(((((.((........)).)))))..)))))))))...........(((((((((.........)))))))))........... (-25.93 = -26.50 + 0.57)

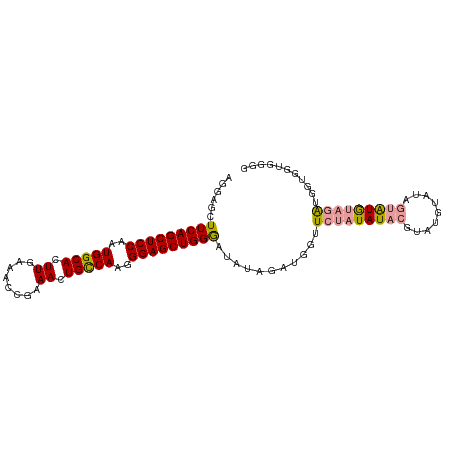
| Location | 10,228,897 – 10,229,000 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.35573 |
| G+C content | 0.49910 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -20.22 |
| Energy contribution | -22.50 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10228897 103 + 22422827 ACAUACGUAUAUAUAACCAUCUAUAUUCCAGCUCCCUUGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCUC--CGGAUUGCUCACUUGAUGGCUUGG ................(((((.......(((((((..(((((.((........)).)))))..))))))).((((((..--.)))..))).....)))))..... ( -31.00, z-score = -2.26, R) >droSim1.chrX_random 589725 103 + 5698898 ACAUACGUAUAUAGAACCAUCUAUAUCCCAGCUCCCUUGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCUC--CGGAUUGCCCACUUGAUGGCUUGG ........(((((((....))))))).((((.(((...((.((((((((((((((......))))))))))))))))..--.)))..(((........))))))) ( -31.80, z-score = -1.98, R) >droSec1.super_15 931833 103 + 1954846 ACAUACGUAUAUAGAACCAUCUAUAUCCCAGCUCCCUUGACAGUUUCGGUUUCAAGUGCCAAUGGAGCUGAAGCUCCUC--CGGAUUGCUCACUUGAUGGCUUGG ........(((((((....)))))))((.((((...((((.....))))..(((((((((...(((((....)))))..--.))......))))))).)))).)) ( -25.00, z-score = -0.45, R) >droEre2.scaffold_4690 13848562 103 - 18748788 AUGUACGUACAUAUAACCAUCUAUAUCACAGCUCCCUCGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCUC--CGGAUUGCUCACUUGAUGGGUUGG ((((....))))....(((((.......(((.(((...((.((((((((((((((......))))))))))))))))..--.)))))).......)))))..... ( -31.54, z-score = -1.79, R) >droAna3.scaffold_13335 838769 82 - 3335858 ----------------------AUGCCACAUGGCCCUCGAUGGCAUCGGUUUCAAGUGGCACUGGAGCUGUAGCUCCUCUCCGGAUUGCUCACUUGAUGGCCCA- ----------------------..((((....(((......))).......((((((((((..(((((....)))))((....)).))).)))))))))))...- ( -29.10, z-score = -0.83, R) >consensus ACAUACGUAUAUAGAACCAUCUAUAUCCCAGCUCCCUUGGCAGUUUCGGUUUCAAGUGCCAUUGGAGCUGAAGCUCCUC__CGGAUUGCUCACUUGAUGGCUUGG ................(((((.......(((.(((...((.((((((((((((((......)))))))))))))))).....)))))).......)))))..... (-20.22 = -22.50 + 2.28)

| Location | 10,228,897 – 10,229,000 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.35573 |
| G+C content | 0.49910 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -25.12 |
| Energy contribution | -27.40 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
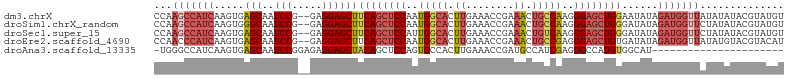
>dm3.chrX 10228897 103 - 22422827 CCAAGCCAUCAAGUGAGCAAUCCG--GAGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCAAGGGAGCUGGAAUAUAGAUGGUUAUAUAUACGUAUGU ...(((((((......((..(((.--..)))))(((((((((..(((((.((........)).)))))..))))))))).....))))))).............. ( -34.80, z-score = -3.15, R) >droSim1.chrX_random 589725 103 - 5698898 CCAAGCCAUCAAGUGGGCAAUCCG--GAGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCAAGGGAGCUGGGAUAUAGAUGGUUCUAUAUACGUAUGU ...(((((((.....(((..(((.--..))))))((((((((..(((((.((........)).)))))..))))))))......))))))).............. ( -33.70, z-score = -1.61, R) >droSec1.super_15 931833 103 - 1954846 CCAAGCCAUCAAGUGAGCAAUCCG--GAGGAGCUUCAGCUCCAUUGGCACUUGAAACCGAAACUGUCAAGGGAGCUGGGAUAUAGAUGGUUCUAUAUACGUAUGU ...(((((((....((((..(((.--..)))))))(((((((.((((((.((........)).)))))).))))))).......))))))).............. ( -32.30, z-score = -1.77, R) >droEre2.scaffold_4690 13848562 103 + 18748788 CCAACCCAUCAAGUGAGCAAUCCG--GAGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCGAGGGAGCUGUGAUAUAGAUGGUUAUAUGUACGUACAU .....(((((....((((..(((.--..)))))))(((((((..(((((.((........)).)))))..))))))).......)))))................ ( -30.70, z-score = -1.80, R) >droAna3.scaffold_13335 838769 82 + 3335858 -UGGGCCAUCAAGUGAGCAAUCCGGAGAGGAGCUACAGCUCCAGUGCCACUUGAAACCGAUGCCAUCGAGGGCCAUGUGGCAU---------------------- -..((((.(((((((.(((.((....))(((((....)))))..))))))))))...((((...))))..)))).........---------------------- ( -30.80, z-score = -1.63, R) >consensus CCAAGCCAUCAAGUGAGCAAUCCG__GAGGAGCUUCAGCUCCAAUGGCACUUGAAACCGAAACUGCCAAGGGAGCUGGGAUAUAGAUGGUUAUAUAUACGUAUGU ...(((((((.....(((..(((.....))))))((((((((..(((((.((........)).)))))..))))))))......))))))).............. (-25.12 = -27.40 + 2.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:51 2011