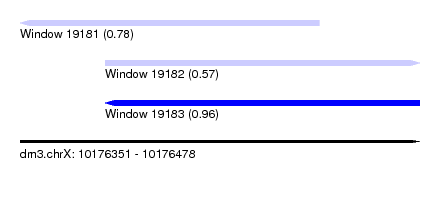
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,176,351 – 10,176,478 |
| Length | 127 |
| Max. P | 0.956084 |

| Location | 10,176,351 – 10,176,446 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 73.29 |
| Shannon entropy | 0.52741 |
| G+C content | 0.51551 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -15.39 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
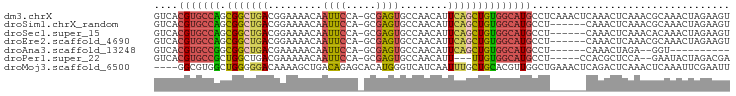
>dm3.chrX 10176351 95 - 22422827 GUCACGUGCCAGCGGCUGACGGAAAACAAUUCCA-GCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAAACUCAAACUCAAACGCAAACUAGAAGU ....(((((((.(((((((.((((.....)))).-((....))......))))))))))))))................................. ( -25.30, z-score = -1.04, R) >droSim1.chrX_random 550623 89 - 5698898 GUCACGUGCCAGCGGCUGACGGAAAACAAUUCCA-GCGAGUGCCAACAUUCAGCUGUGGCAUGCCU------CAAACUCAAACGCAAACUAGAAGU ....(((((((.(((((((.((((.....)))).-((....))......))))))))))))))...------........................ ( -25.30, z-score = -1.09, R) >droSec1.super_15 879188 89 - 1954846 GUCACGUGCCAGCGGCUGACGGAAAACAAUUCCA-GCGAGUGCCAACAUUCAGCUGUGGCAUGCCU------CAAACUCAAACACAAACUAGAAGU ....(((((((.(((((((.((((.....)))).-((....))......))))))))))))))...------........................ ( -25.30, z-score = -1.51, R) >droEre2.scaffold_4690 13796138 89 + 18748788 GUCACGUGCCAGCGGCUGACGGAAAACAAUUCCA-GCGAGUGCCAACAUUCAGCUGUGGCAUGCCU------CAAACUCAAACGCAAACUAGAAGU ....(((((((.(((((((.((((.....)))).-((....))......))))))))))))))...------........................ ( -25.30, z-score = -1.09, R) >droAna3.scaffold_13248 1271652 77 + 4840945 GUCACGUGCCGGCGGCUGACGAAAAACAAUUCCA-GCGAGUGCCAACAUUCAGCUGUGGCAUGCCU------CAAACUAGA--GGU---------- ....(((((((.(((((((.........((((..-..))))........))))))))))))))(((------(......))--)).---------- ( -26.63, z-score = -1.76, R) >droPer1.super_22 326317 85 + 1688296 GUCACGUGCCGCUGGCUGACGAAAAACAAUUCCA-GCGAGUGCCAACAUU---UUGUGGCAUGCCU-----CCACGCUCCA--GAAUACUAGACGA (((..(..(((((((.((........))...)))-))).)..)....(((---(((..((.((...-----.)).))..))--))))....))).. ( -24.20, z-score = -1.37, R) >droMoj3.scaffold_6500 24884087 92 + 32352404 ----GGCGUGGCUGGGGGACAAAAGCUGACAGAGCACAUGGGUCAUCAAUUUGCUGCACGUUGGCUGAAACUCAGACUCAAACUCAAAUUCGAAUU ----...((((((.(....)...)))).)).(((....((((((........(((.......)))((.....))))))))..)))........... ( -18.40, z-score = 1.20, R) >consensus GUCACGUGCCAGCGGCUGACGGAAAACAAUUCCA_GCGAGUGCCAACAUUCAGCUGUGGCAUGCCU______CAAACUCAAACGCAAACUAGAAGU ....(((((((.(((((((.........((((.....))))........))))))))))))))................................. (-15.39 = -15.60 + 0.21)
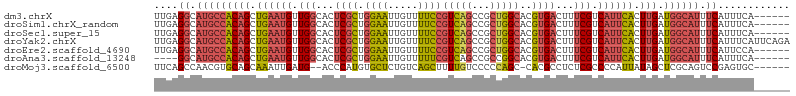
| Location | 10,176,378 – 10,176,478 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.37064 |
| G+C content | 0.50259 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10176378 100 + 22422827 UUGAGGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUCCGUCAGCCGCUGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUUCA------ ....((.(((((((((.((((((.(((.(.((((.((((.....))))(((((...)))))..)))).).))).)))))).))).)))))).))......------ ( -31.60, z-score = -1.36, R) >droSim1.chrX_random 550644 100 + 5698898 UUGAGGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUCCGUCAGCCGCUGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUUCA------ ....((.(((((((((.((((((.(((.(.((((.((((.....))))(((((...)))))..)))).).))).)))))).))).)))))).))......------ ( -31.60, z-score = -1.36, R) >droSec1.super_15 879209 100 + 1954846 UUGAGGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUCCGUCAGCCGCUGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUUCA------ ....((.(((((((((.((((((.(((.(.((((.((((.....))))(((((...)))))..)))).).))).)))))).))).)))))).))......------ ( -31.60, z-score = -1.36, R) >droYak2.chrX 18749555 106 + 21770863 UUGAGGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUCCGUCAGCCGCUGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUUCAUUCAGA ....((.(((((((((.((((((.(((.(.((((.((((.....))))(((((...)))))..)))).).))).)))))).))).)))))).))............ ( -31.60, z-score = -0.92, R) >droEre2.scaffold_4690 13796159 100 - 18748788 UUGAGGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUCCGUCAGCCGCUGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUCCA------ ....((.(((((((((.((((((.(((.(.((((.((((.....))))(((((...)))))..)))).).))).)))))).))).)))))).))......------ ( -31.60, z-score = -1.22, R) >droAna3.scaffold_13248 1271665 96 - 4840945 ----GGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUUCGUCAGCCGCCGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUUCA------ ----((.(((((((((.(((.(((((((....(((((((......)))..)))).))))))).(((((....)))))))).))).)))))).))......------ ( -31.10, z-score = -1.88, R) >droMoj3.scaffold_6500 24884114 97 - 32352404 UUCAGCCAACGUGCAGCAAAUUGAUG--ACCCAUGUGCUCUGUCAGCUUUUGUCCCCCAGC-CACGCCUCUCGCCCCAUUAUAGCUCGCAGUCCGAGUGC------ ...(((..(((.(.((((...((...--...))..))))))))..))).............-.....................(((((.....)))))..------ ( -13.30, z-score = 2.19, R) >consensus UUGAGGCAUGCCACAGCUGAAUGUUGGCACUCGCUGGAAUUGUUUUCCGUCAGCCGCUGGCACGUGACUUUCGUCAUUCACUUGAUGGCAUUUCAUUUCA______ ....((.(((((((((.((((((.(((.(.((((.((((.....))))(((((...)))))..)))).).))).)))))).))).)))))).))............ (-23.58 = -24.10 + 0.52)

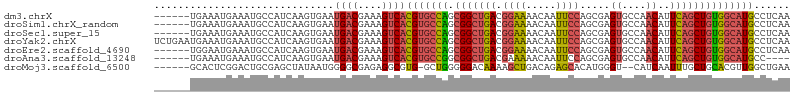
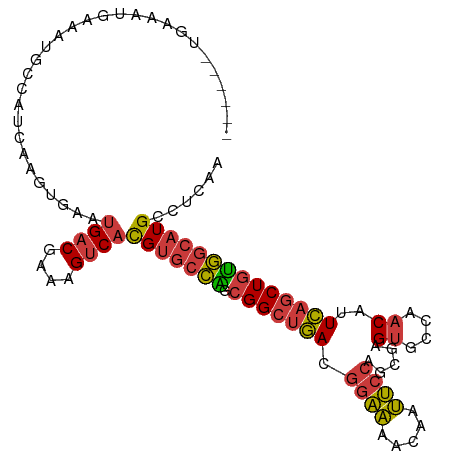
| Location | 10,176,378 – 10,176,478 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.37064 |
| G+C content | 0.50259 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10176378 100 - 22422827 ------UGAAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCAGCGGCUGACGGAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAA ------.....(((................((((....))))(((((((.(((((((.((((.....)))).((....))......))))))))))))))..))). ( -32.80, z-score = -2.36, R) >droSim1.chrX_random 550644 100 - 5698898 ------UGAAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCAGCGGCUGACGGAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAA ------.....(((................((((....))))(((((((.(((((((.((((.....)))).((....))......))))))))))))))..))). ( -32.80, z-score = -2.36, R) >droSec1.super_15 879209 100 - 1954846 ------UGAAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCAGCGGCUGACGGAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAA ------.....(((................((((....))))(((((((.(((((((.((((.....)))).((....))......))))))))))))))..))). ( -32.80, z-score = -2.36, R) >droYak2.chrX 18749555 106 - 21770863 UCUGAAUGAAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCAGCGGCUGACGGAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAA ...........(((................((((....))))(((((((.(((((((.((((.....)))).((....))......))))))))))))))..))). ( -32.80, z-score = -1.95, R) >droEre2.scaffold_4690 13796159 100 + 18748788 ------UGGAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCAGCGGCUGACGGAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAA ------.((.....................((((....))))(((((((.(((((((.((((.....)))).((....))......)))))))))))))))).... ( -33.10, z-score = -2.03, R) >droAna3.scaffold_13248 1271665 96 + 4840945 ------UGAAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCGGCGGCUGACGAAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCC---- ------.........((((((....(((..((((....))))..)))((((.((((..(((......)))))))(((((....))))).))))))))))...---- ( -30.20, z-score = -2.01, R) >droMoj3.scaffold_6500 24884114 97 + 32352404 ------GCACUCGGACUGCGAGCUAUAAUGGGGCGAGAGGCGUG-GCUGGGGGACAAAAGCUGACAGAGCACAUGGGU--CAUCAAUUUGCUGCACGUUGGCUGAA ------(((.......))).(((((...((.((((((..(.(((-(((.(....)....(((.....))).....)))--))))..)))))).))...)))))... ( -26.20, z-score = 0.83, R) >consensus ______UGAAAUGAAAUGCCAUCAAGUGAAUGACGAAAGUCACGUGCCAGCGGCUGACGGAAAACAAUUCCAGCGAGUGCCAACAUUCAGCUGUGGCAUGCCUCAA ..............................((((....))))(((((((.(((((((.((((.....)))).....((....))..))))))))))))))...... (-25.15 = -25.30 + 0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:44 2011