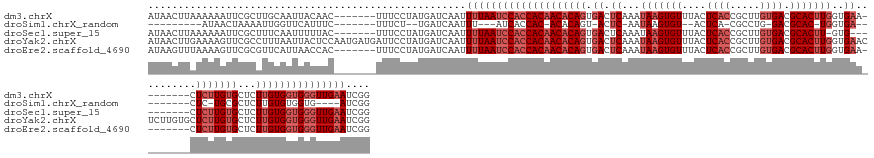
| Sequence ID | dm3.chrX |
|---|---|
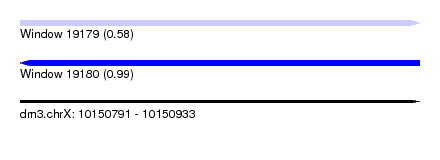
| Location | 10,150,791 – 10,150,933 |
| Length | 142 |
| Max. P | 0.989014 |

| Location | 10,150,791 – 10,150,933 |
|---|---|
| Length | 142 |
| Sequences | 5 |
| Columns | 157 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Shannon entropy | 0.36418 |
| G+C content | 0.40432 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.48 |
| Covariance contribution | 1.49 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
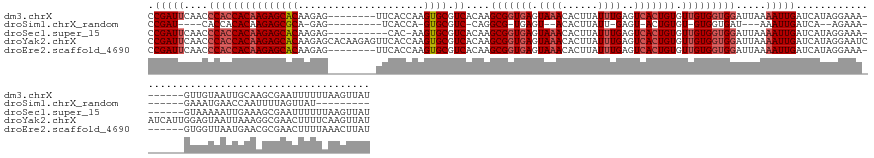
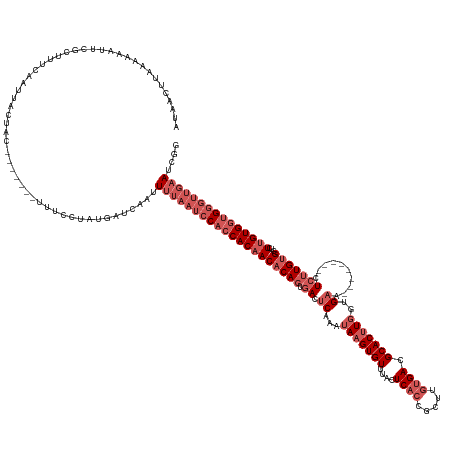
>dm3.chrX 10150791 142 + 22422827 CCGAUUCAACCCACCACAAGAGCACAAGAG--------UUCACCAAGUGCGUCACAAGCGGUGAGUAAACACUUAUUUGAGUCACUGUGUUGUGGUGGAUUAAAAUUGAUCAUAGGAAA-------GUUGUAAUUGCAAGCGAAUUUUUUAAGUUAU .(((((....(((((((((((((((.....--------........)))).))....(((((((.((((......))))..))))))).))))))))).....)))))....(((((((-------.((((........)))).)))))))...... ( -34.52, z-score = -1.29, R) >droSim1.chrX_random 531790 114 + 5698898 CCGAU----CACCACACAAGAGCGCA-GAG---------UCACCA-GUGCGUC-CAGGCG-UGAGU--ACACUUAUU-GAGU-ACUGUGU-GUGGUGAU---AAAUUGAUCA--AGAAA-------GAAAUGAACCAAUUUUAGUUAU--------- ...((----(((((((((.(..((((-..(---------....).-.))))..-).....-..(((--((.(.....-).))-))).)))-))))))))---((((((.(((--.....-------....)))..)))))).......--------- ( -29.70, z-score = -1.89, R) >droSec1.super_15 862519 139 + 1954846 CCGAUUCAACCCACCACAAGAGCACAAGAG----------CAC-AAGUGCGUCACAAGCGGUGAGUAAACACUUAUUUGAGUCACUGUGUUGUGGUGGAUUAAAAUUGAUCAUAGGAAA-------GUAAAAAUUGAAAGCGAAUUUUUUAAGUUAU .(((((....(((((((((((((((.....----------...-..)))).))....(((((((.((((......))))..))))))).))))))))).....)))))...........-------((((...(((((((.....))))))).)))) ( -32.00, z-score = -1.35, R) >droYak2.chrX 18732698 157 + 21770863 CCGAUUCAACCCACCACAAGAGCACAAGAGCACAAGAGUUCACCAAGUGCGUCACAAGCGGUGAGUAAACACUUAUUUGAGUCACUGUGUUGUGGUGGAUUAAAAUUGAUCAUAGGAAUCAUCAUUGGAGUAAUUAAAGGCGAACUUUUCAAGUUAU ..(((((....((((((((((((((..((((......)))).....)))).))....(((((((.((((......))))..))))))).))))))))(((((....)))))....))))).........((((((((((.....))))...)))))) ( -39.00, z-score = -1.37, R) >droEre2.scaffold_4690 13778607 142 - 18748788 CCGAUUCAACCCACCACAAGAGCACAAGAG--------UUCACCAAGUGCGUCACAAGCGGUGAGUAAACACUUAUUUGAGUCACUGUGUUGUGGUGGAUUAAAAUUGAUCAUAGGAAA-------GUGGUUAAUGAACGCGAACUUUUAAACUUAU .((.(((...(((((((((((((((.....--------........)))).))....(((((((.((((......))))..))))))).)))))))))......(((((((((......-------))))))))))))))................. ( -37.12, z-score = -1.56, R) >consensus CCGAUUCAACCCACCACAAGAGCACAAGAG________UUCACCAAGUGCGUCACAAGCGGUGAGUAAACACUUAUUUGAGUCACUGUGUUGUGGUGGAUUAAAAUUGAUCAUAGGAAA_______GUAGUAAUUGAAAGCGAAUUUUUUAAGUUAU .(((((....(((((((((((((((.....................)))).))....(((((((.((((......))))..))))))).))))))))).....)))))................................................. (-21.99 = -23.48 + 1.49)
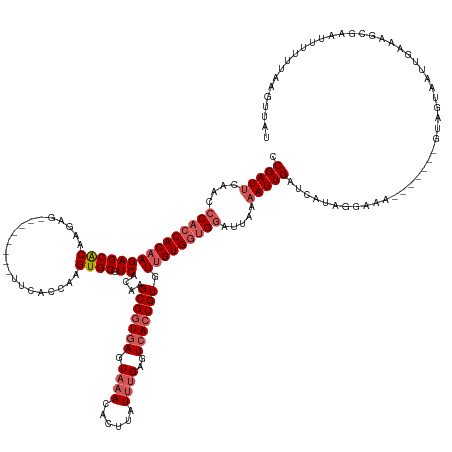
| Location | 10,150,791 – 10,150,933 |
|---|---|
| Length | 142 |
| Sequences | 5 |
| Columns | 157 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Shannon entropy | 0.36418 |
| G+C content | 0.40432 |
| Mean single sequence MFE | -36.36 |
| Consensus MFE | -21.52 |
| Energy contribution | -23.72 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10150791 142 - 22422827 AUAACUUAAAAAAUUCGCUUGCAAUUACAAC-------UUUCCUAUGAUCAAUUUUAAUCCACCACAACACAGUGACUCAAAUAAGUGUUUACUCACCGCUUGUGACGCACUUGGUGAA--------CUCUUGUGCUCUUGUGGUGGGUUGAAUCGG ...............................-------...............((((((((((((((((((((.((.(((..(((((((....((((.....)))).))))))).))).--------.)))))))...))))))))))))))).... ( -35.10, z-score = -2.56, R) >droSim1.chrX_random 531790 114 - 5698898 ---------AUAACUAAAAUUGGUUCAUUUC-------UUUCU--UGAUCAAUUU---AUCACCAC-ACACAGU-ACUC-AAUAAGUGU--ACUCA-CGCCUG-GACGCAC-UGGUGA---------CUC-UGCGCUCUUGUGUGGUG----AUCGG ---------.......((((((((.((....-------.....--))))))))))---((((((((-(((.(((-((.(-.....).))--)))..-.....(-..((((.-.(....---------)..-))))..).)))))))))----))... ( -35.30, z-score = -4.09, R) >droSec1.super_15 862519 139 - 1954846 AUAACUUAAAAAAUUCGCUUUCAAUUUUUAC-------UUUCCUAUGAUCAAUUUUAAUCCACCACAACACAGUGACUCAAAUAAGUGUUUACUCACCGCUUGUGACGCACUU-GUG----------CUCUUGUGCUCUUGUGGUGGGUUGAAUCGG ........(((((((.......)))))))..-------...............((((((((((((((((((((.((.....((((((((....((((.....)))).))))))-)).----------.)))))))...))))))))))))))).... ( -35.20, z-score = -3.53, R) >droYak2.chrX 18732698 157 - 21770863 AUAACUUGAAAAGUUCGCCUUUAAUUACUCCAAUGAUGAUUCCUAUGAUCAAUUUUAAUCCACCACAACACAGUGACUCAAAUAAGUGUUUACUCACCGCUUGUGACGCACUUGGUGAACUCUUGUGCUCUUGUGCUCUUGUGGUGGGUUGAAUCGG .............................((..((((.((....)).))))..((((((((((((((((((((.((.....(((((.(((((((....((.......))....))))))).)))))..)))))))...)))))))))))))))..)) ( -40.50, z-score = -2.38, R) >droEre2.scaffold_4690 13778607 142 + 18748788 AUAAGUUUAAAAGUUCGCGUUCAUUAACCAC-------UUUCCUAUGAUCAAUUUUAAUCCACCACAACACAGUGACUCAAAUAAGUGUUUACUCACCGCUUGUGACGCACUUGGUGAA--------CUCUUGUGCUCUUGUGGUGGGUUGAAUCGG ..................(.((((.......-------......)))).)...((((((((((((((((((((.((.(((..(((((((....((((.....)))).))))))).))).--------.)))))))...))))))))))))))).... ( -35.72, z-score = -1.73, R) >consensus AUAACUUAAAAAAUUCGCUUUCAAUUACUAC_______UUUCCUAUGAUCAAUUUUAAUCCACCACAACACAGUGACUCAAAUAAGUGUUUACUCACCGCUUGUGACGCACUUGGUGAA________CUCUUGUGCUCUUGUGGUGGGUUGAAUCGG .....................................................((((((((((((((((((((((((.(......).)).)))((((((..((.....))..)))))).............))))...))))))))))))))).... (-21.52 = -23.72 + 2.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:41 2011