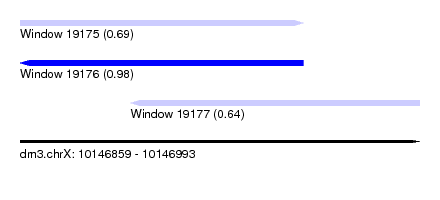
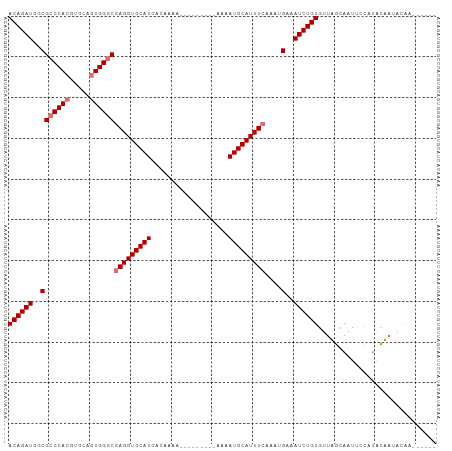
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,146,859 – 10,146,993 |
| Length | 134 |
| Max. P | 0.983875 |

| Location | 10,146,859 – 10,146,954 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Shannon entropy | 0.32047 |
| G+C content | 0.41847 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
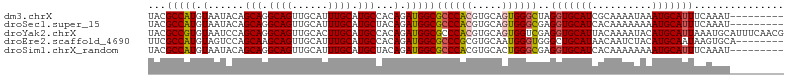
>dm3.chrX 10146859 95 + 22422827 UUCUGAAUUUCUUGC-UGGAAUUGCAAACACAGAUUUCAUUUGAAAUGCAUUUA---------UUUUGCGAUGCACCUAGCCCACUGCACGUGGGCGCCAUCUGU ...((((((((....-.)))))).))...((((((..(........((((((..---------......))))))....((((((.....)))))))..)))))) ( -25.20, z-score = -1.46, R) >droSec1.super_15 858623 90 + 1954846 ------UUGUAUAGUAUGGAAUUGCUAACACAGAUUUCAUUUGAAAUGCAUUUU---------UUUUGUGAUGCACCUCGCCCACUGCACGUGGGCGCCAUCUGU ------..((.(((((......)))))))((((((..(....((..((((((..---------......))))))..))((((((.....)))))))..)))))) ( -27.80, z-score = -2.60, R) >droYak2.chrX 18728187 93 + 21770863 ------------UACAAAUAAUUACAAACACAGAUUUCCGUUGAAAUGCAUUUAAUGCAUGUAUUUUGUAAUGCACCUCGACCACUGCACGUGGGCGCCAUCUGU ------------.................((((((..(((((((..((((((.((((....))))....))))))..)))))(((.....)))))....)))))) ( -19.30, z-score = -0.52, R) >droSim1.chrX_random 527494 90 + 5698898 ------UUGUAUAGUAUGGAAUUGCUAACACAGAUUUCAUUUGAAAUGCAUUUU---------UUUUGUGAUGCACCUCGCCCAGUGCACGUGGGCGCCAUCUGU ------..((.(((((......)))))))((((((..(....((..((((((..---------......))))))..))((((((....).))))))..)))))) ( -24.40, z-score = -1.13, R) >consensus ______UUGUAUAGCAUGGAAUUGCAAACACAGAUUUCAUUUGAAAUGCAUUUA_________UUUUGUGAUGCACCUCGCCCACUGCACGUGGGCGCCAUCUGU ...........(((((......)))))..((((((...........((((((.................))))))....((((((.....))))))...)))))) (-18.12 = -18.80 + 0.69)

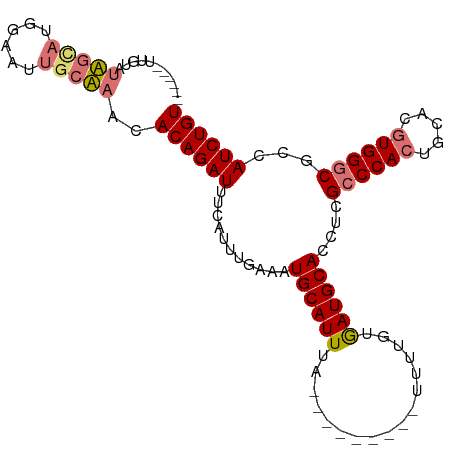
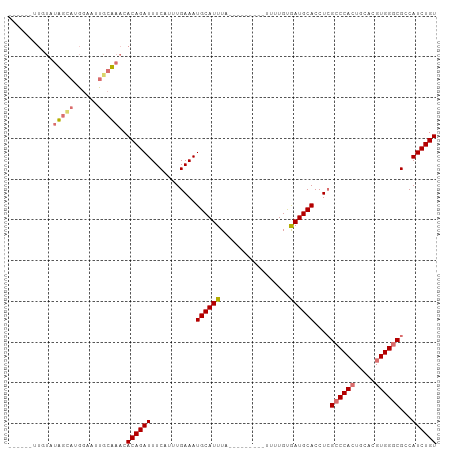
| Location | 10,146,859 – 10,146,954 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Shannon entropy | 0.32047 |
| G+C content | 0.41847 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.91 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10146859 95 - 22422827 ACAGAUGGCGCCCACGUGCAGUGGGCUAGGUGCAUCGCAAAA---------UAAAUGCAUUUCAAAUGAAAUCUGUGUUUGCAAUUCCA-GCAAGAAAUUCAGAA ((((((..(((((((.....)))))).((((((((.......---------...)))))))).....)..)))))).(((((.......-))))).......... ( -27.70, z-score = -1.52, R) >droSec1.super_15 858623 90 - 1954846 ACAGAUGGCGCCCACGUGCAGUGGGCGAGGUGCAUCACAAAA---------AAAAUGCAUUUCAAAUGAAAUCUGUGUUAGCAAUUCCAUACUAUACAA------ ((((((..(((((((.....))))))(((((((((.......---------...)))))))))....)..)))))).......................------ ( -29.70, z-score = -3.38, R) >droYak2.chrX 18728187 93 - 21770863 ACAGAUGGCGCCCACGUGCAGUGGUCGAGGUGCAUUACAAAAUACAUGCAUUAAAUGCAUUUCAACGGAAAUCUGUGUUUGUAAUUAUUUGUA------------ (((((((((((((((.....))).....)))))((((((((....((((((...))))))....((((....)))).))))))))))))))).------------ ( -25.00, z-score = -0.99, R) >droSim1.chrX_random 527494 90 - 5698898 ACAGAUGGCGCCCACGUGCACUGGGCGAGGUGCAUCACAAAA---------AAAAUGCAUUUCAAAUGAAAUCUGUGUUAGCAAUUCCAUACUAUACAA------ ((((((..((((((.(....))))))(((((((((.......---------...)))))))))....)..)))))).......................------ ( -25.50, z-score = -2.15, R) >consensus ACAGAUGGCGCCCACGUGCAGUGGGCGAGGUGCAUCACAAAA_________AAAAUGCAUUUCAAAUGAAAUCUGUGUUAGCAAUUCCAUACAAUACAA______ ((((((..(((((((.....))))))(((((((((...................)))))))))....)..))))))............................. (-24.16 = -24.91 + 0.75)

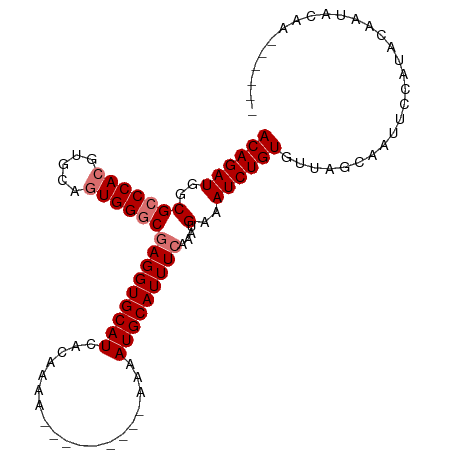
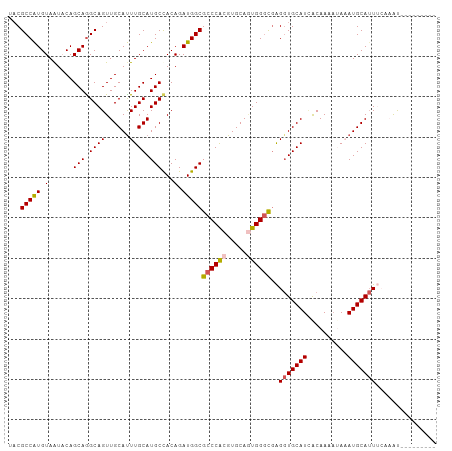
| Location | 10,146,896 – 10,146,993 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.28092 |
| G+C content | 0.49068 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
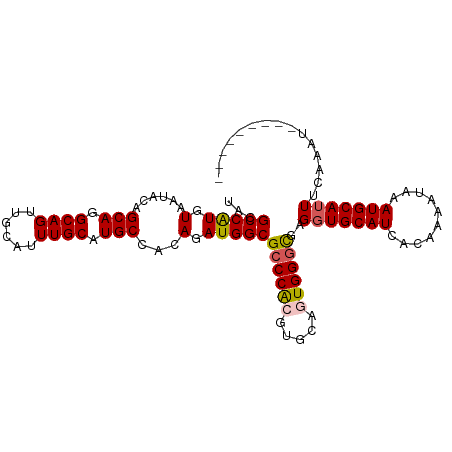
>dm3.chrX 10146896 97 - 22422827 UACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCCACAGAUGGCGCCCACGUGCAGUGGGCUAGGUGCAUCGCAAAAUAAAUGCAUUUCAAAU--------- ...(((.(((......))))))..((((...(((((((((....))))((((((.....))))))...)))))..))))..................--------- ( -33.70, z-score = -1.37, R) >droSec1.super_15 858655 97 - 1954846 UACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCUACAGAUGGCGCCCACGUGCAGUGGGCGAGGUGCAUCACAAAAAAAAUGCAUUUCAAAU--------- ...(((((.(.....((((.((((......)))).))))..).)))))((((((.....))))))(((((((((..........)))))))))....--------- ( -37.70, z-score = -3.32, R) >droYak2.chrX 18728213 106 - 21770863 UACGCCGUGUAAUCCAGCAGGCAGUUGCACUUGCAUGCCACAGAUGGCGCCCACGUGCAGUGGUCGAGGUGCAUUACAAAAUACAUGCAUUAAAUGCAUUUCAACG ((((((.(((......))))))...(((((.((..(((((....)))))..)).))))))))((.((((((((((.................)))))))))).)). ( -32.63, z-score = -0.55, R) >droEre2.scaffold_4690 13774557 98 + 18748788 UUCGCCAUGUAGUCCAGCAAGCAGUUGCAUUUGCAUGCCACAGAUGGCGCCCGCGUGCAAUGGGUGGGCUGCAUAACAAUCUACAUGCAAUAAGUGCA-------- ......((((((((((.(.....(((((((.((..(((((....)))))..)).))))))).).))))))))))...........((((.....))))-------- ( -33.40, z-score = 0.12, R) >droSim1.chrX_random 527526 97 - 5698898 UACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCUACAGAUGGCGCCCACGUGCACUGGGCGAGGUGCAUCACAAAAAAAAUGCAUUUCAAAU--------- ...(((((.(.....((((.((((......)))).))))..).)))))(((((.(....))))))(((((((((..........)))))))))....--------- ( -33.50, z-score = -2.02, R) >consensus UACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCCACAGAUGGCGCCCACGUGCAGUGGGCGAGGUGCAUCACAAAAUAAAUGCAUUUCAAAU_________ ...(((((.(......(((.((((......)))).)))...).)))))((((((.....))))))..(((((((..........)))))))............... (-26.50 = -26.82 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:39 2011