| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,140,085 – 10,140,245 |
| Length | 160 |
| Max. P | 0.975678 |

| Location | 10,140,085 – 10,140,245 |
|---|---|
| Length | 160 |
| Sequences | 4 |
| Columns | 178 |
| Reading direction | forward |
| Mean pairwise identity | 69.25 |
| Shannon entropy | 0.48148 |
| G+C content | 0.44103 |
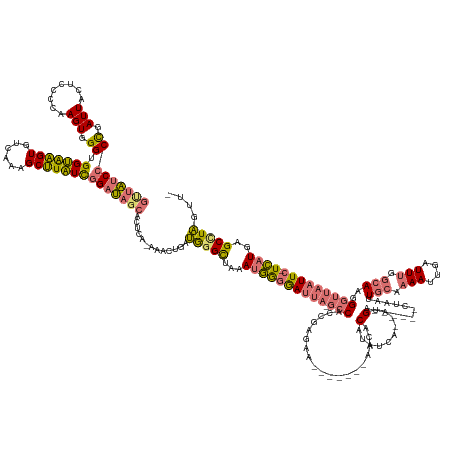
| Mean single sequence MFE | -52.88 |
| Consensus MFE | -23.50 |
| Energy contribution | -20.82 |
| Covariance contribution | -2.69 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
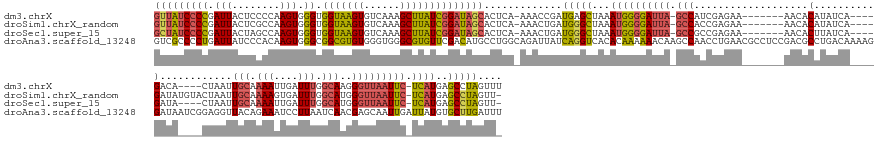
>dm3.chrX 10140085 160 + 22422827 GUUAUCCCCGAUUACUCCCCAAGUGGGUGGUAAGUGUCAAAGCUUAUCGGAUAGCACUCA-AAACCGAUGAGCUAAAUGGGGAUUA-GCCAUCGAGAA-------AACACAUAUCA----GACA----CUAAUUGCAAAAUUGAUUUGGCAAGGGUUAAUUC-UCAUGAGCCUAGUUU ...(((((((.((((..(((....)))..)))).......((((((((((..........-...))))))))))...)))))))..-(((((.(((((-------(((.....(((----((..----.(((((....))))).))))).....)))..)))-))))).))....... ( -45.42, z-score = -1.99, R) >droSim1.chrX_random 519204 163 + 5698898 GUUAUCCCCGAUUACUCGCCAAGUGGGUGGUAAGUGUCAAAGCUUAUCGGAUAGCACUCA-AAACUGAUGGGCUAAAUGGGGAUUA-GCCACCGAGAA-------AACACAUAUCA----GAUAUGUACUAAUUGCAAAAGUGAUUUGGCAUGGGUUAAUUC-UCAUGAGCCUAGUU- (((((((......((((((...))))))(((((((......)))))))))))))).....-.......((((((..((((((((((-(((...((...-------........)).----..(((((...(((..(....)..)))..))))))))))))))-)))).))))))...- ( -52.40, z-score = -2.97, R) >droSec1.super_15 850294 159 + 1954846 GCUAUCCCCGAUUACUAGCCAAGUGGGUGGUAAGUGUCAAAGCUUAUCGGAUAGCACUCA-AAACUGAUGGGCUAAAUGGGGAUUA-GCCGCCGAGAA-------AACACUUAUCA----GAUA----CUAAUUGCAAAAUUGAUUUGGCAUGGGUUAAUUC-UCAUGAGCCUAGUU- (((((((((.(((........))).)).(((((((......)))))))))))))).....-.......((((((..((((((((((-(((((((((..-------....((....)----)...----.(((((....))))).))))))...)))))))))-)))).))))))...- ( -50.40, z-score = -2.57, R) >droAna3.scaffold_13248 1227318 178 - 4840945 GUCGCCCCUGAUUAUCCCACAAGUGGGCGGCGUGUGGGUGGGCGUGUUCGACAUGCCUGGCAGAUUAUCAGGUCACACAAAAAACAAGCCAACCUGAACGCCUCCGACGCCUGACAAAAGGAUAAUCGGAGGUUACAGAAAUCCUUAAUCAACGAGCAAUUGAUUAUGUGCUUGAUUU (((((((((............)).)))))))((((((.((((((((.....)))))))).).(.....)....)))))......(((((((..(((...((((((((..(((......)))....))))))))..))).......(((((((.......))))))))).))))).... ( -63.30, z-score = -2.93, R) >consensus GUUAUCCCCGAUUACUCCCCAAGUGGGUGGUAAGUGUCAAAGCUUAUCGGAUAGCACUCA_AAACUGAUGGGCUAAAUGGGGAUUA_GCCACCGAGAA_______AACACAUAUCA____GAUA____CUAAUUGCAAAAUUGAUUUGGCAAGGGUUAAUUC_UCAUGAGCCUAGUU_ ..(((((((.(((........)))))).))))..((((((((((((((((..............))))))))))...(((........)))......................................................)))))).((.(((........))).))...... (-23.50 = -20.82 + -2.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:36 2011