| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,811,510 – 10,811,607 |
| Length | 97 |
| Max. P | 0.980861 |

| Location | 10,811,510 – 10,811,607 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.55884 |
| G+C content | 0.53229 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.33 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
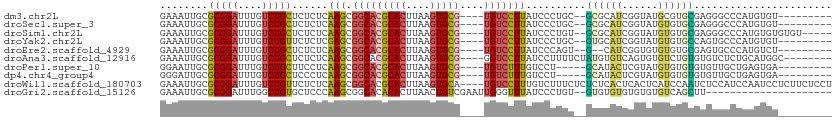
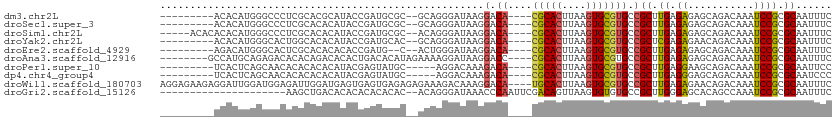
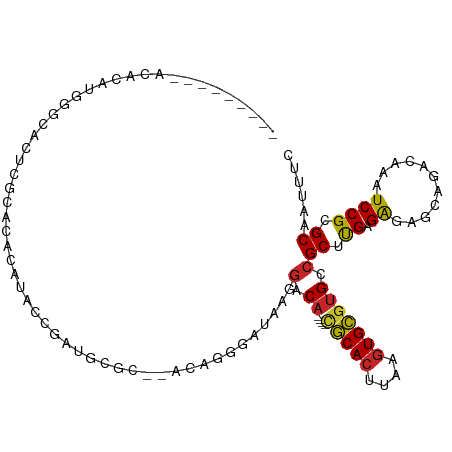
>dm3.chr2L 10811510 97 + 23011544 GAAAUUGCGCGGAUUUGUCUGCUCUCUCAAGCGGCACGCACUUAAGUGCG----UGUCCUUAUCCCUGC--GCGCAUCGGUAUGCGUGCGAGGGCCCAUGUGU--------- ......(((((((....)))((((((..(((.(((((((((....)))))----)))))))......((--((((((....))))))))))))))....))))--------- ( -42.30, z-score = -2.93, R) >droSec1.super_3 6229928 97 + 7220098 GAAAUUGCGCGGAUUUGUCUGCUCUCUCAAGCGGCACGCACUUAAGUGCG----UGUCCUUAUCCCUGC--GCGCAUCGGUAUGUGUGCGAGGGCCCAUGUGU--------- ......(((((((....)))((((((..(((.(((((((((....)))))----)))))))......((--((((((....))))))))))))))....))))--------- ( -40.40, z-score = -2.67, R) >droSim1.chr2L 10617906 101 + 22036055 GAAAUUGCGCGGAUUUGUCUGCUCUCUCAAGCGGCACGCACUUAAGUGCG----UGUCCUUAUCCCUGU--GCGCAUCGGUAUGUGUGCGAGGGCCCAUGUGUGUGU----- ......(((((.((.((...((((((..(((.(((((((((....)))))----)))))))......(.--.(((((....)))))..))))))).)).)).)))))----- ( -41.00, z-score = -2.74, R) >droYak2.chr2L 7218532 97 + 22324452 GAAAUUGCGCGGAUUUGUCUGUUCUCUCGAGCGGCACGCACUUAAGUGCG----UGUCCUUAUCCCUGC--GUGCAUCGGUAUGUGUGCCAGUGCCCAUGUGU--------- ......((((((........((((....))))(((((((((....)))))----)))).......))))--))((((.((((....)))).))))........--------- ( -36.00, z-score = -2.47, R) >droEre2.scaffold_4929 12022371 95 - 26641161 GAAAUUGCGCGGAUUUGUCUGCUCUCUCAAGCGGCACGCACUUAAGUGCG----UGUCCUUAUCCCAGU--G--CAUCGGUGUGUGUGCGAGUGCCCAUGUCU--------- .....(((((((((..(...(((......)))(((((((((....)))))----)))))..))))..))--)--))..((..((.(.((....)))))..)).--------- ( -32.40, z-score = -1.47, R) >droAna3.scaffold_12916 14662869 100 + 16180835 GAAAUUGCGCGGAUUUGUCUGCUCUCUCAAGCGGCACGCACUUAAGUGCG----GGUCCUUAUCCUUUUCUAUGUGUCAGUGUGUCUGUGUGUCUCUGCAUGGC-------- ......((((((((....(((((......)))(((((((((....))))(----(((....))))........)))))))...))))))))(((.......)))-------- ( -27.50, z-score = -0.68, R) >droPer1.super_10 2030643 94 + 3432795 GGAAUUGCGCGGAUUUGUCUGCUUCCUCAAGCGGCACGCACUUAAGUGCG----UGUCUUUGUCCU-----GCAUACUCGUAUGUGUGUGUGUUGCUGAGUGA--------- ........(((((....))))).((((((.(((((((((((....(((((----.((...((....-----.)).)).)))))..))))))))))))))).))--------- ( -35.80, z-score = -2.96, R) >dp4.chr4_group4 3015992 94 + 6586962 GGGAUUGCGCGGAUUUGUCUGCUCCCUCAAGCGGCACGCACUUAAGUGCG----UGUCUUUGUCCU-----GCAUACUCGUAUGUGUGUGUGUUGCUGAGUGA--------- ((((....(((((....)))))))))(((.(((((((((((....(((((----.((...((....-----.)).)).)))))..))))))))))))))....--------- ( -37.10, z-score = -2.88, R) >droWil1.scaffold_180703 369708 108 - 3946847 GAAAUUGCGCGGAUUUGUCUGUUCUCUCAAGCGGCACGCACUUAAGUGCA----UGUCCUUUGUCUUUCUCUCUCACUCACUCAUCCAAUCUCCAUCCAAUCCUCUUCUCCU ((.((((.(((((....))))).....((((.((((.((((....)))).----)))).)))).......................)))).))................... ( -17.00, z-score = -1.61, R) >droGri2.scaffold_15126 2041137 89 + 8399593 GAAAUUGCGCGGAUUUGGCUGUGCUCCCAAGCGGCACACACUUAACUGUCGAAUUGGGUUUAUCCCUGU--GUGUGUGUGUGUGUCAGCUU--------------------- ......((((((......))))))....(((((((((((((...((...((....(((.....)))..)--).))..))))))))).))))--------------------- ( -31.30, z-score = -2.10, R) >consensus GAAAUUGCGCGGAUUUGUCUGCUCUCUCAAGCGGCACGCACUUAAGUGCG____UGUCCUUAUCCCUGC__GCGCAUCGGUAUGUGUGCGAGUGCCCAUGUGU_________ ..........((((.((.(((((......)))))))(((((....)))))...........))))......((((((......))))))....................... (-15.42 = -15.33 + -0.09)
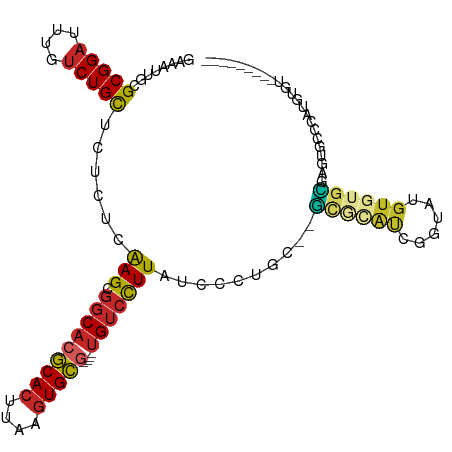
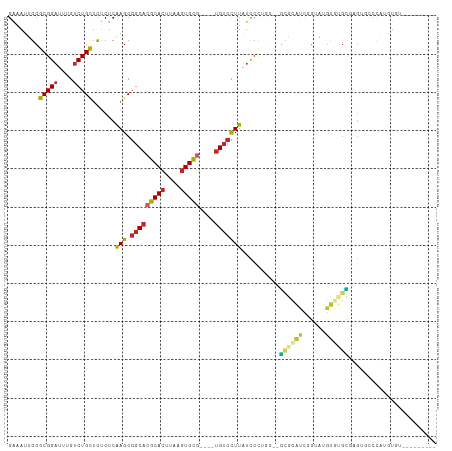
| Location | 10,811,510 – 10,811,607 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.55884 |
| G+C content | 0.53229 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -8.45 |
| Energy contribution | -8.47 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10811510 97 - 23011544 ---------ACACAUGGGCCCUCGCACGCAUACCGAUGCGC--GCAGGGAUAAGGACA----CGCACUUAAGUGCGUGCCGCUUGAGAGAGCAGACAAAUCCGCGCAAUUUC ---------.....((.((....((.(((((....))))).--))..((((..((.((----(((((....)))))))))((((....))))......)))))).))..... ( -40.30, z-score = -3.82, R) >droSec1.super_3 6229928 97 - 7220098 ---------ACACAUGGGCCCUCGCACACAUACCGAUGCGC--GCAGGGAUAAGGACA----CGCACUUAAGUGCGUGCCGCUUGAGAGAGCAGACAAAUCCGCGCAAUUUC ---------.....((.((.(((((.((........)).))--).))((((..((.((----(((((....)))))))))((((....))))......)))))).))..... ( -35.60, z-score = -2.94, R) >droSim1.chr2L 10617906 101 - 22036055 -----ACACACACAUGGGCCCUCGCACACAUACCGAUGCGC--ACAGGGAUAAGGACA----CGCACUUAAGUGCGUGCCGCUUGAGAGAGCAGACAAAUCCGCGCAAUUUC -----........(((.((....))...))).....(((((--....((((..((.((----(((((....)))))))))((((....))))......)))))))))..... ( -34.70, z-score = -3.33, R) >droYak2.chr2L 7218532 97 - 22324452 ---------ACACAUGGGCACUGGCACACAUACCGAUGCAC--GCAGGGAUAAGGACA----CGCACUUAAGUGCGUGCCGCUCGAGAGAACAGACAAAUCCGCGCAAUUUC ---------........(((.(((........))).)))..--((..((((..((.((----(((((....)))))))))(.((....)).)......))))..))...... ( -29.60, z-score = -2.11, R) >droEre2.scaffold_4929 12022371 95 + 26641161 ---------AGACAUGGGCACUCGCACACACACCGAUG--C--ACUGGGAUAAGGACA----CGCACUUAAGUGCGUGCCGCUUGAGAGAGCAGACAAAUCCGCGCAAUUUC ---------.....((.((....((((.......).))--)--....((((..((.((----(((((....)))))))))((((....))))......)))))).))..... ( -32.90, z-score = -3.01, R) >droAna3.scaffold_12916 14662869 100 - 16180835 --------GCCAUGCAGAGACACACAGACACACUGACACAUAGAAAAGGAUAAGGACC----CGCACUUAAGUGCGUGCCGCUUGAGAGAGCAGACAAAUCCGCGCAAUUUC --------....(((.........(((.....)))............((((..((.(.----(((((....))))).)))((((....))))......))))..)))..... ( -26.40, z-score = -2.95, R) >droPer1.super_10 2030643 94 - 3432795 ---------UCACUCAGCAACACACACACAUACGAGUAUGC-----AGGACAAAGACA----CGCACUUAAGUGCGUGCCGCUUGAGGAAGCAGACAAAUCCGCGCAAUUCC ---------........................((((.(((-----.(((....(.((----(((((....))))))).)((((....)))).......)))..))))))). ( -24.40, z-score = -2.01, R) >dp4.chr4_group4 3015992 94 - 6586962 ---------UCACUCAGCAACACACACACAUACGAGUAUGC-----AGGACAAAGACA----CGCACUUAAGUGCGUGCCGCUUGAGGGAGCAGACAAAUCCGCGCAAUCCC ---------..((((..................)))).(((-----.(((....(.((----(((((....))))))).)((((....)))).......)))..)))..... ( -23.97, z-score = -1.44, R) >droWil1.scaffold_180703 369708 108 + 3946847 AGGAGAAGAGGAUUGGAUGGAGAUUGGAUGAGUGAGUGAGAGAGAAAGACAAAGGACA----UGCACUUAAGUGCGUGCCGCUUGAGAGAACAGACAAAUCCGCGCAAUUUC .........((((((..((((..(((..((......(....).......(((.((.((----(((((....)))))))))..)))......))..))).))))..)))))). ( -25.10, z-score = -1.89, R) >droGri2.scaffold_15126 2041137 89 - 8399593 ---------------------AAGCUGACACACACACACAC--ACAGGGAUAAACCCAAUUCGACAGUUAAGUGUGUGCCGCUUGGGAGCACAGCCAAAUCCGCGCAAUUUC ---------------------..((((.........(((((--((.(((.....)))(((......)))..)))))))..(((....))).))))................. ( -21.10, z-score = -0.66, R) >consensus _________ACACAUGGGCACUCGCACACAUACCGAUGCGC__ACAGGGAUAAGGACA____CGCACUUAAGUGCGUGCCGCUUGAGAGAGCAGACAAAUCCGCGCAAUUUC ...............................................................((((....))))((((.(.(((..........)))..).))))...... ( -8.45 = -8.47 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:04 2011