
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,125,239 – 10,125,336 |
| Length | 97 |
| Max. P | 0.883536 |

| Location | 10,125,239 – 10,125,336 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 58.16 |
| Shannon entropy | 0.71436 |
| G+C content | 0.69160 |
| Mean single sequence MFE | -53.34 |
| Consensus MFE | -17.27 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.90 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10125239 97 - 22422827 ------------------GGUGGCGGUGCCAUUUCCGGUGGCUGGAGC---UCCGGCGGAGGUGCCGCUUCCGGUGGCUGGAGCUCCGGCGGAGGUGCCG---CUUCCGGUGGCUGGAGCA ------------------((.(((((..((...(((....((((((((---(((((((((((.....)))))....)))))))))))))))))))..)))---)).))....((....)). ( -57.10, z-score = -2.39, R) >droYak2.chrU 18185677 97 - 28119190 ------------------GGUGGUGGUGCCAUCUCCGGUGGCUGGAGC---GCCGGAGGCGGUGGCUCAUCCGGCGGCUGGAGUGCCGGAGGUGGUGCCC---GGAAUUGUGGCUGGAGCU ------------------....(.((..((((((((((((.(..(..(---(((((((((....)))..))))))).)..)..))))))))))))..)))---........(((....))) ( -51.40, z-score = -2.33, R) >droEre2.scaffold_4690 13750925 97 + 18748788 ------------------GGUGGAGGUGCCAUUUCCGGUGGCUGGAGU---GCCGGAGGAGGUGGCUCGUCCGGUGGCUGGAGCGCCGGAGGAGGUGCCC---CGUCCGGUGGCUGGAGCA ------------------(((((.((..((.(((((((((.(..(..(---((((((.(((....))).))))))).)..)..))))))))).))..)))---)).))....((....)). ( -52.60, z-score = -2.70, R) >droAna3.scaffold_13047 940716 115 - 1816235 GGCGGCGGUGCCUCUUCCAAUAGCGGCGGAGGCACCAGCGGCUGGAGC---UCCGGUGGCGCCUCCCCCUCGGGAGGCUGGAGCUCAAGCGGCUCCUCCU---CGUCGGGGGGCUGGAGCU (((((((((((((((.((......)).))))))))).((.(((((...---.))))).)))))..((((((((((((..((((((.....)))))).)))---).)))))))).....))) ( -71.70, z-score = -2.44, R) >dp4.Unknown_singleton_3695 20824 103 - 24995 ------------------AAUGGUGGUUCCUCCCAUGGGGGUGGCGGCUGGAGCAGCGGUGGUUCUGCUCCCAGCAGUGGCUGGAGCAGUGGUGGCUCCGUUGCCAGCAGCGGCUGGAGCA ------------------.......(((((.((....))(((.((.((((..(((((((.(((((((((((.(((....))))))))))....))))))))))))))).)).)))))))). ( -51.10, z-score = -0.82, R) >droPer1.super_13 332624 103 + 2293547 ------------------AAUGGUGGUUCCUCCCAUGGGGGUGGCGGCUGGAGCAGCGGUGGUUCCGCUCCCAGCAGUGGCUGGAGCAGUGGUGGCUCCGUUGCCAGCAGCGGCUGGAGCA ------------------.......(((((.((....))(((.((.((((..(((((((.(((((((((((((((....))))).).))))).))))))))))))))).)).)))))))). ( -52.00, z-score = -0.96, R) >droWil1.scaffold_181096 2064515 97 + 12416693 ------------------AAUGGUGGCUCGGCAUCUGCGGGUUGGAGC---AAUAACGGUGGCUCCUCCUCUGGUGGUUGGAGCAAUAACGGUGGCUCCU---CCUCUGGUGGUUGGAGCA ------------------.......(((((.((((...(((..(((((---....((.((.(((((.((......))..)))))....)).)).))))).---)))..)))).)..)))). ( -37.50, z-score = -0.94, R) >consensus __________________AAUGGUGGUGCCACCUCCGGGGGCUGGAGC___ACCGGCGGUGGUUCCGCUUCCGGCGGCUGGAGCACCAGCGGUGGCUCCG___CCUCCGGUGGCUGGAGCA ......................((((...((((.((....(((((.......))))))).))))))))(((((((.((.(((........((.....))......))).)).))))))).. (-17.27 = -16.57 + -0.70)
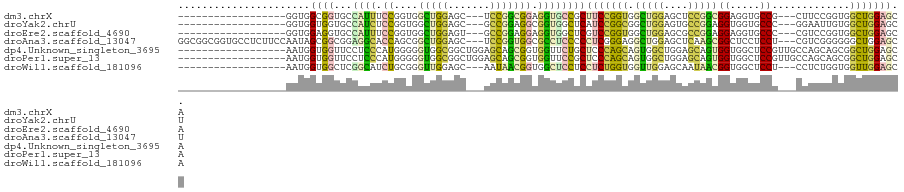
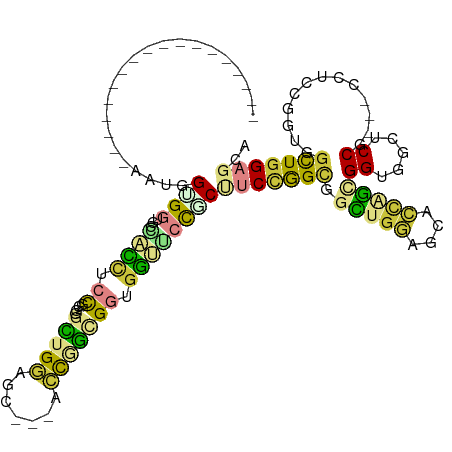
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:35 2011