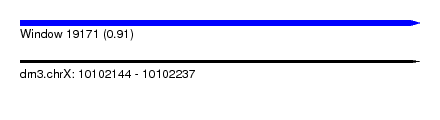
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,102,144 – 10,102,237 |
| Length | 93 |
| Max. P | 0.914861 |

| Location | 10,102,144 – 10,102,237 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 61.73 |
| Shannon entropy | 0.77242 |
| G+C content | 0.51183 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -10.73 |
| Energy contribution | -10.66 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10102144 93 + 22422827 ---AUGCAGUAGCAACAGCGGUAAAAACUGCCGCGACUGCUGUCAUUGAACAAUGAUUUCUGCGUUUAAAG---UCACUUGCCUCGCGCGAUUUUUGAU ---((((((.((((...((((((.....))))))...))))((((((....))))))..))))))((((((---....((((.....)))).)))))). ( -25.70, z-score = -0.78, R) >droPer1.super_56 36002 84 + 431378 CUGCAGCGGCAAAUGCAGCAGCAGAAACUGCUGCGGCUGCUGUCAUUGAACGAUUACAGAGGCGUCCCUGUCCCGCCACUGAUA--------------- ..((((((((....(((((((......))))))).)))))))).............(((.((((.........)))).)))...--------------- ( -34.60, z-score = -2.49, R) >dp4.chrXL_group3b 17635 84 + 386183 CUGCAGCGGCAAAUGCAGCAGCAGAAACUGCUGCGGCUGCUGUCAUUGAACGAUUACAGAGGCGUCCCUGUCCCGCCACUGAUA--------------- ..((((((((....(((((((......))))))).)))))))).............(((.((((.........)))).)))...--------------- ( -34.60, z-score = -2.49, R) >droSim1.chrX_random 480582 93 + 5698898 ---ACGCAGUAGCAGCAGCGGUAAAACCUGCCGCGGCUGCUGUCAUUGAACAAUGAUUUCUGCGUUUCAAG---UCACUUGCCGCGUGCGAUUUUUGAU ---((((((.((((((.((((((.....)))))).))))))((((((....))))))..)))))).(((((---....((((.....))))..))))). ( -35.70, z-score = -2.51, R) >droSec1.super_15 812637 93 + 1954846 ---ACGCAGUAGCAGCAGCGGUAAAACCUGCCGCGGCUGCUGUCAUUGAACACUGAUUUCUGCGUUUCAAG---UCACUUGCCGCGUGCGAUUUUUGAU ---((((((.((((((.((((((.....)))))).))))))((((........))))..)))))).(((((---....((((.....))))..))))). ( -32.80, z-score = -1.47, R) >droYak2.chrX 18678975 93 + 21770863 ---AAGCAGAAGCAGCAGCGGUAAAAACGGCCGCGGCUGCUGUCAUUGAACAAUGAUUUCUGCGAUUACAG---UCACUUGCUUCGUGCGAUUCUGAAU ---..(((((((((((.(((((.......))))).))))))((((((....)))))).))))).....(((---....((((.....))))..)))... ( -35.90, z-score = -3.02, R) >droEre2.scaffold_4690 13728301 93 - 18748788 ---AAGCAAAAGCAGCAGCGGUAAAAACUGCCGCGGCUGCUGUCAUUGAAUAAUGAUUUCUGCGAUUACAG---UCACUUGCCGCGCGCGAUUCUUAAU ---..((...((((((.((((((.....)))))).))))))((((((....))))))....(((....(((---....)))...))))).......... ( -28.60, z-score = -0.94, R) >droWil1.scaffold_180702 2262186 95 - 4511350 ---AGACAACAGAAGCAACAGCAAAUGCUGCUGCGGCCGCUGUCAUUGAACGGUUUAUAUUGGGCUUACAGCUCUCACUUGCUCUUGACCAAUUGCAG- ---.((((.(.(..(((.((((....)))).)))..).).))))........((....(((((......(((........))).....))))).))..- ( -23.30, z-score = 0.49, R) >droAna3.scaffold_13047 905945 71 + 1816235 -----------GCGGCGUCGGUGGAGACUGGCUCGGCUGCUGUCAUUGAGUUAUAAUUGCUGCGAAUUCAG---UGGACUCCUCU-------------- -----------(((((((((((....))))))...)))))..((((((((((............)))))))---)))........-------------- ( -20.50, z-score = 0.17, R) >consensus ___AAGCAGCAGCAGCAGCGGUAAAAACUGCCGCGGCUGCUGUCAUUGAACAAUGAUUUCUGCGUUUACAG___UCACUUGCUGCG_GCGAUUUUU_AU ..............((.((((((.....))))))(((....))).................)).................................... (-10.73 = -10.66 + -0.07)

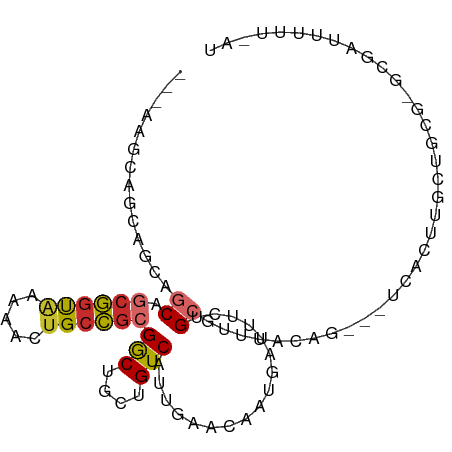
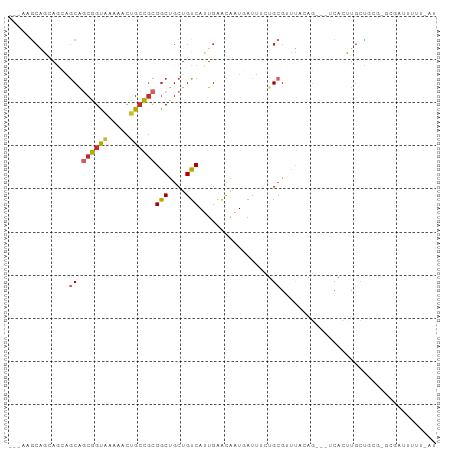
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:34 2011