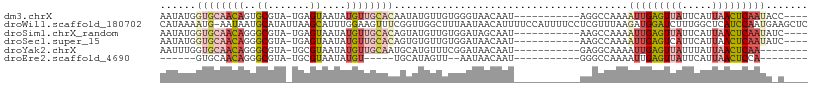
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,076,157 – 10,076,249 |
| Length | 92 |
| Max. P | 0.995419 |

| Location | 10,076,157 – 10,076,249 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.59 |
| Shannon entropy | 0.60969 |
| G+C content | 0.35273 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -7.48 |
| Energy contribution | -9.48 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
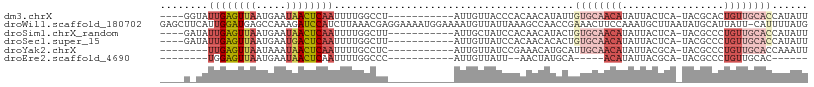
>dm3.chrX 10076157 92 + 22422827 ----GGUAUUGAGUUAAUGAAUAACUCAAUUUUGGCCU-----------AUUGUUACCCACAACAUAUUGUGCAACAUAUUACUCA-UACGCACUGUUGCACCAUAUU ----((((((((((((.....)))))))))....))).-----------...............((((.((((((((.........-.......)))))))).)))). ( -20.69, z-score = -2.24, R) >droWil1.scaffold_180702 2227632 107 - 4511350 GAGCUUCAUUGGAUGAGCCAAAGAUCCAUCUUAAACGAGGAAAAUGGAAAAUGUUAUUAAAGCCAACCGAAACUUCCAAAUGCUUAAUAUGCAUUAUU-CAUUUUAUG ...((((..(((.....)))((((....))))....))))((((((((.(((((((((((.((.....((....)).....)))))))).))))).))-))))))... ( -18.80, z-score = -0.81, R) >droSim1.chrX_random 2873751 92 + 5698898 ----GAUAUUGAGUUAAUGAAUAACUCAAUUUUGGCUU-----------AUUGCUAUCCACAACAUACUGUGCAACAUAUUACUCA-UACGCCCUGUUGCACCAUAUU ----...(((((((((.....)))))))))..((((..-----------...)))).............((((((((.........-.......))))))))...... ( -19.79, z-score = -2.96, R) >droSec1.super_15 786949 92 + 1954846 ----GAUAUUGAGUUAAUGAAUGACUCAAUUUUGGCUU-----------AUUGUUAUCCACAACACACUGUGCAACAUAUUACUCA-UACGCCCUGUUGCACCAUAUU ----...(((((((((.....)))))))))..((((..-----------...)))).............((((((((.........-.......))))))))...... ( -17.39, z-score = -1.56, R) >droYak2.chrX 18652089 88 + 21770863 --------UUGAGUUAAUAAAUAACUCAAUUUUGCCUC-----------AUUGUUAUCCGAAACAUGCAUUGCAACAUAUUACGCA-UACGCCCUGUUGCACCAAAUU --------((((((((.....))))))))...(((...-----------..((((......)))).))).(((((((......((.-...))..)))))))....... ( -16.10, z-score = -2.01, R) >droEre2.scaffold_4690 13702595 75 - 18748788 --------UGGAGUUAAUGAAUAACUCAAUUUUGGCCC-----------AUUGUUAUU--AACUAUGCA-----ACAUAUUACGCA-UACGCCCUGUUGCAC------ --------...(((((((((.....(((....)))...-----------....)))))--)))).((((-----(((......((.-...))..))))))).------ ( -13.10, z-score = -0.43, R) >consensus ____G_UAUUGAGUUAAUGAAUAACUCAAUUUUGGCUU___________AUUGUUAUCCAAAACAUACUGUGCAACAUAUUACUCA_UACGCCCUGUUGCACCAUAUU ........((((((((.....))))))))........................................((((((((.................))))))))...... ( -7.48 = -9.48 + 2.00)
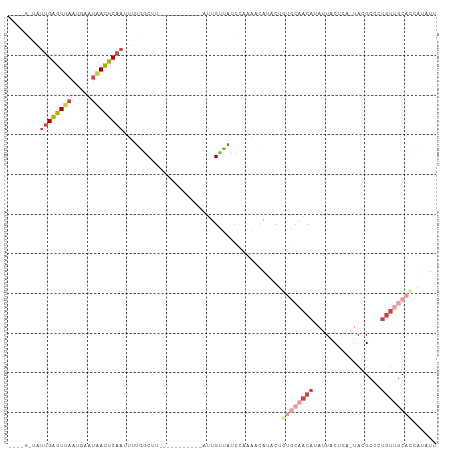
| Location | 10,076,157 – 10,076,249 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 65.59 |
| Shannon entropy | 0.60969 |
| G+C content | 0.35273 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -12.06 |
| Energy contribution | -13.87 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
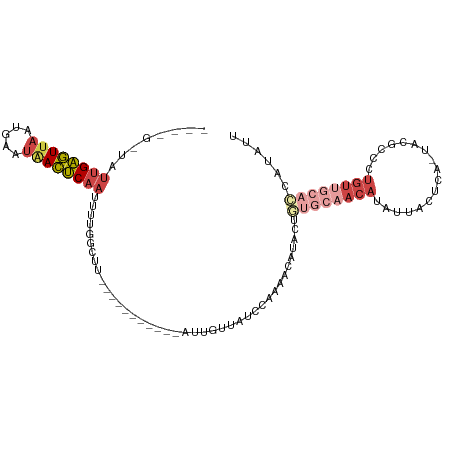
>dm3.chrX 10076157 92 - 22422827 AAUAUGGUGCAACAGUGCGUA-UGAGUAAUAUGUUGCACAAUAUGUUGUGGGUAACAAU-----------AGGCCAAAAUUGAGUUAUUCAUUAACUCAAUACC---- ....((((((((((.(((...-...)))...))))))......((((((.....)))))-----------).))))..(((((((((.....)))))))))...---- ( -28.30, z-score = -3.22, R) >droWil1.scaffold_180702 2227632 107 + 4511350 CAUAAAAUG-AAUAAUGCAUAUUAAGCAUUUGGAAGUUUCGGUUGGCUUUAAUAACAUUUUCCAUUUUCCUCGUUUAAGAUGGAUCUUUGGCUCAUCCAAUGAAGCUC .........-...(((((.......)))))....(((((((.((((.........((...((((((((........))))))))....))......))))))))))). ( -18.76, z-score = 0.14, R) >droSim1.chrX_random 2873751 92 - 5698898 AAUAUGGUGCAACAGGGCGUA-UGAGUAAUAUGUUGCACAGUAUGUUGUGGAUAGCAAU-----------AAGCCAAAAUUGAGUUAUUCAUUAACUCAAUAUC---- ......((((((((..((...-...))....)))))))).((.((((((.....)))))-----------).))....(((((((((.....)))))))))...---- ( -27.70, z-score = -3.53, R) >droSec1.super_15 786949 92 - 1954846 AAUAUGGUGCAACAGGGCGUA-UGAGUAAUAUGUUGCACAGUGUGUUGUGGAUAACAAU-----------AAGCCAAAAUUGAGUCAUUCAUUAACUCAAUAUC---- ......((((((((..((...-...))....)))))))).((.((((((.....)))))-----------).))....(((((((.........)))))))...---- ( -24.10, z-score = -2.03, R) >droYak2.chrX 18652089 88 - 21770863 AAUUUGGUGCAACAGGGCGUA-UGCGUAAUAUGUUGCAAUGCAUGUUUCGGAUAACAAU-----------GAGGCAAAAUUGAGUUAUUUAUUAACUCAA-------- .......(((((((..(((..-..)))....))))))).....(((((((........)-----------))))))...((((((((.....))))))))-------- ( -25.20, z-score = -2.80, R) >droEre2.scaffold_4690 13702595 75 + 18748788 ------GUGCAACAGGGCGUA-UGCGUAAUAUGU-----UGCAUAGUU--AAUAACAAU-----------GGGCCAAAAUUGAGUUAUUCAUUAACUCCA-------- ------((((((((..(((..-..)))....)))-----)))))....--.........-----------...........((((((.....))))))..-------- ( -18.90, z-score = -1.50, R) >consensus AAUAUGGUGCAACAGGGCGUA_UGAGUAAUAUGUUGCACAGUAUGUUGUGGAUAACAAU___________AAGCCAAAAUUGAGUUAUUCAUUAACUCAAUA_C____ ......((((((((..((.......))....)))))))).......................................(((((((((.....)))))))))....... (-12.06 = -13.87 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:29 2011