| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,072,459 – 10,072,567 |
| Length | 108 |
| Max. P | 0.973828 |

| Location | 10,072,459 – 10,072,567 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.41029 |
| G+C content | 0.44833 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -16.51 |
| Energy contribution | -14.35 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
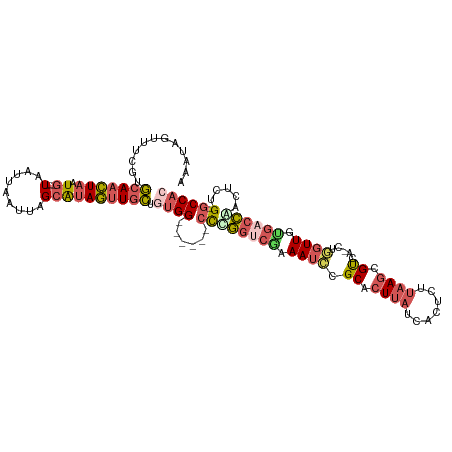
>dm3.chrX 10072459 108 + 22422827 GUGGC-CAGAGUUGGUCACAACCAG-UGACGCUUAAGAGUGAUAAGUGCGGAUUUCGACCGGG------CCACAGCAACUAUGCUAAUUAAUUA---CAUUAGUUGCACGAAACUAUUU (((((-(.(.(((((((((.....)-))))(((((.......)))))........))))).))------)))).(((((((((.(((....)))---)).)))))))............ ( -33.60, z-score = -2.92, R) >droSec1.super_15 783192 108 + 1954846 GUGGC-CAGAGUCGGUCACAACCAG-UGGCGCUUAAGAGUGAUAAGUGCGGAUUUCGACCGGG------CCACAGCAACUAUGCUAAUUAAUUA---CAUUAGUUGCACAGAACUAUUU (((((-(.(.(((((.(((.....)-))(((((((.......))))))).....)))))).))------)))).(((((((((.(((....)))---)).)))))))............ ( -37.20, z-score = -3.60, R) >droYak2.chrX 18648147 108 + 21770863 GUGGC-CAGAGUUGGUCACAACGAG-UGACGCUUAAGAGUGAUAAGUGCGGAUUUUGGCCAGG------CCACAGCAACUAUGCUAAUUAAUUA---CAUUAGUUGCACGAAACUAUUU .((((-(((((((.(((((.....)-))))(((((.......)))))...)))))))))))..------.....(((((((((.(((....)))---)).)))))))............ ( -32.70, z-score = -2.45, R) >droEre2.scaffold_4690 13698604 108 - 18748788 GUGGC-CAGAGUCGGUCACAACCAG-UGACGCUUAAGAGUGAUAAGUGCGGAUUUAGACCAGG------CCAGCGCAACUAUGCUAAUUAAUUA---CAUUAGUUGCACGGAACUAUUU .((((-(.(.(((.(((((.....)-))))(((((.......))))).........)))).))------)))(.(((((((((.(((....)))---)).))))))).).......... ( -30.10, z-score = -1.59, R) >droAna3.scaffold_13047 881112 119 + 1816235 GUGGUGCAAACUUCGGGCCAAGGAGACUGCUUUUGAGCAAGGAGACUGCUCUUUCGGGGGGAGAGCUCCCCGAAAGAGUUGCGCAAAUUAAUUAGUUUAUUAAAUUUGAGAAACUAUUU ...(((((..((((..((..(((((....)))))..))..))))...((((((((((((((....)))))))))))))))))))........((((((.(((....))).))))))... ( -44.60, z-score = -3.78, R) >consensus GUGGC_CAGAGUUGGUCACAACCAG_UGACGCUUAAGAGUGAUAAGUGCGGAUUUCGACCGGG______CCACAGCAACUAUGCUAAUUAAUUA___CAUUAGUUGCACGAAACUAUUU ((((.......((((((....((...((.((((....)))).)).....)).....)))))).......)))).(((((((...................)))))))............ (-16.51 = -14.35 + -2.16)


| Location | 10,072,459 – 10,072,567 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.41029 |
| G+C content | 0.44833 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -11.18 |
| Energy contribution | -12.86 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10072459 108 - 22422827 AAAUAGUUUCGUGCAACUAAUG---UAAUUAAUUAGCAUAGUUGCUGUGG------CCCGGUCGAAAUCCGCACUUAUCACUCUUAAGCGUCA-CUGGUUGUGACCAACUCUG-GCCAC ............(((((((.((---(.........)))))))))).((((------((.(((((.((((.((.((((.......)))).))..-..)))).)))))......)-))))) ( -28.60, z-score = -1.74, R) >droSec1.super_15 783192 108 - 1954846 AAAUAGUUCUGUGCAACUAAUG---UAAUUAAUUAGCAUAGUUGCUGUGG------CCCGGUCGAAAUCCGCACUUAUCACUCUUAAGCGCCA-CUGGUUGUGACCGACUCUG-GCCAC ............(((((((.((---(.........)))))))))).((((------((((((((.((((.((.((((.......)))).))..-..)))).)))))).....)-))))) ( -33.30, z-score = -2.84, R) >droYak2.chrX 18648147 108 - 21770863 AAAUAGUUUCGUGCAACUAAUG---UAAUUAAUUAGCAUAGUUGCUGUGG------CCUGGCCAAAAUCCGCACUUAUCACUCUUAAGCGUCA-CUCGUUGUGACCAACUCUG-GCCAC .....((..((.(((((((.((---(.........))))))))))))..)------).((((((.........((((.......)))).((((-(.....)))))......))-)))). ( -28.00, z-score = -2.14, R) >droEre2.scaffold_4690 13698604 108 + 18748788 AAAUAGUUCCGUGCAACUAAUG---UAAUUAAUUAGCAUAGUUGCGCUGG------CCUGGUCUAAAUCCGCACUUAUCACUCUUAAGCGUCA-CUGGUUGUGACCGACUCUG-GCCAC ..........(((((((((.((---(.........))))))))))))(((------((.((((..........((((.......)))).((((-(.....))))).))))..)-)))). ( -33.90, z-score = -3.35, R) >droAna3.scaffold_13047 881112 119 - 1816235 AAAUAGUUUCUCAAAUUUAAUAAACUAAUUAAUUUGCGCAACUCUUUCGGGGAGCUCUCCCCCCGAAAGAGCAGUCUCCUUGCUCAAAAGCAGUCUCCUUGGCCCGAAGUUUGCACCAC .....(((.(.((((((.(((......))))))))).).)))((((((((((........))))))))))((((.((.(.((((....))))(((.....)))..).)).))))..... ( -30.10, z-score = -2.38, R) >consensus AAAUAGUUUCGUGCAACUAAUG___UAAUUAAUUAGCAUAGUUGCUGUGG______CCCGGUCGAAAUCCGCACUUAUCACUCUUAAGCGUCA_CUGGUUGUGACCAACUCUG_GCCAC ............(((((((...................))))))).((((........((((((.((((.((.((((.......)))).)).....)))).))))))........)))) (-11.18 = -12.86 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:27 2011