| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,062,206 – 10,062,302 |
| Length | 96 |
| Max. P | 0.826389 |

| Location | 10,062,206 – 10,062,302 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.40 |
| Shannon entropy | 0.60321 |
| G+C content | 0.33904 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -9.02 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10062206 96 + 22422827 -------------------AUAUAUGAAUUGUAAUCACUUUGCCUGUGAUUUUUGU-CAUAAUUGCAAU-CAAAUCUCAAAAACUCAAUCUUUAGCAAAGUUUGUGUGCUCAACUUU -------------------...(((((.....((((((.......))))))....)-))))...(((..-(((((........................)))))..)))........ ( -12.86, z-score = -0.05, R) >droSim1.chrX_random 447543 116 + 5698898 AAAUAUACAUAAAUUCCUCUUACAUAUAUUGUAAUCACCUUGCCUGUGAUUUUUGUUCAUAAUUGCAGU-CAAAUCUCAAAAACUCAAUCUUGGGCAAAGUUUUUGGGCUCAACUUU ...................(((((.....))))).......(((((.((.(((((((((..((((.(((-............)))))))..))))))))).)).)))))........ ( -20.20, z-score = -1.86, R) >droSec1.super_15 773498 116 + 1954846 AAAUAUACAUAAAUUCCUCUUACAUAUAUUGUAAUCACCUUGCCUGUGAUUUUUGUUCAUAAUUGCAGU-CAAAUCUCAAAAACUCAAUCUUUGGCAAAGUUUUUGGGCUCAACUUU ...................(((((.....))))).....(((.(((..(((.........)))..))).-)))..((((((((((.............))))))))))......... ( -17.22, z-score = -1.18, R) >droYak2.chrX 18637162 109 + 21770863 ------AAAUAUAAAUACCUCUUCCAAAUUGUAAUCACUUUGCCUGUGAUUUUUGU-CAUAAUUGCAGU-CAAAUCUCAAAAACUCAAUCUUUAGCAAAGUUUUUUGGCUCAACUUU ------.................(((((........(((((((.((((((....))-)))).(((.((.-.....)))))..............)))))))..)))))......... ( -13.80, z-score = -1.06, R) >droEre2.scaffold_4690 13688651 103 - 18748788 ------------AAAUUCCUCUUUCAAAUUGUAAUCACUUUGUCUGUGAUUUUUGU-CAUAAUUGCAGU-CAGAUCUCAAAAACUCAAUCGUUGGCAAAGUUUUUGGGCUCAACUUU ------------............................((.(((..(((.....-...)))..))).-))((.((((((((((.............)))))))))).))...... ( -18.82, z-score = -1.57, R) >droAna3.scaffold_13047 872800 73 + 1816235 -----------------------------------------GGCGCCGGGCACUUUGUGUGUUUUCAGCAUAAAUCUCUGGAACUCGGUC-UGGGGAAAGUUUCUGGGG--AACUUU -----------------------------------------..(.(((((.((((..(((((.....)))))..((((..((......))-..)))))))).))))).)--...... ( -20.70, z-score = -0.68, R) >consensus ____________A_____CUUAUAUAAAUUGUAAUCACUUUGCCUGUGAUUUUUGU_CAUAAUUGCAGU_CAAAUCUCAAAAACUCAAUCUUUGGCAAAGUUUUUGGGCUCAACUUU ...........................................((((((((.........)))))))).......((((((((((.............))))))))))......... ( -9.02 = -9.30 + 0.28)

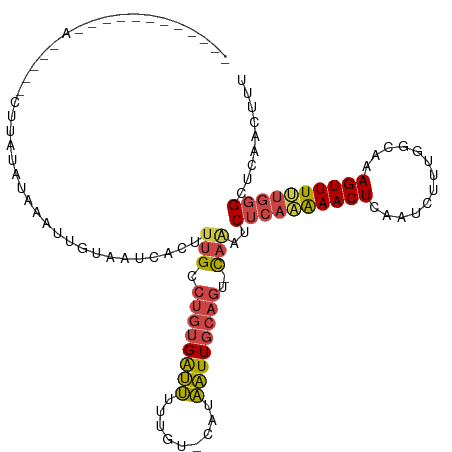

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:23 2011