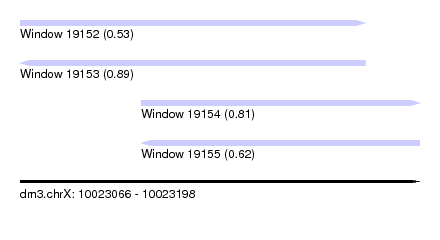
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,023,066 – 10,023,198 |
| Length | 132 |
| Max. P | 0.886203 |

| Location | 10,023,066 – 10,023,180 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.41 |
| Shannon entropy | 0.25463 |
| G+C content | 0.42150 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
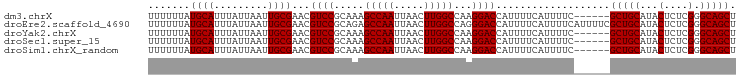
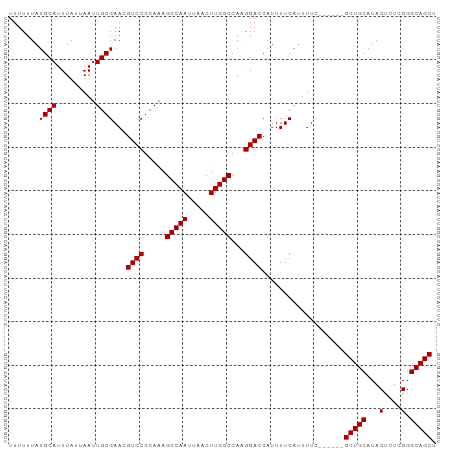
>dm3.chrX 10023066 114 + 22422827 CGUUCGGCGGAUGUAGUGUUCUACUCGUUUCCCGAGAUCGUUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUCGCU------ .((((.((((((((.........((((.....)))).((((..((.(((....))).))..))))))))))))...(((((.....)))))...))))................------ ( -29.70, z-score = -1.87, R) >droEre2.scaffold_4690 13656140 104 - 18748788 ----------------CGUUUUUCUCGUUAACCGAGAUCGUUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAGAGCCAAUUAACUUGGCCAGGGACCAUUUUCAUUUUCAUUUUCGCU ----------------.((((.(((((.....)))))...........(((.........)))))))((((.(.(.(((((.....)))))).)))))...................... ( -19.90, z-score = -0.83, R) >droYak2.chrX 18604177 96 + 21770863 ----------------CGUUC--CUCGUUAACCGAGAUCGUUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUCGCU------ ----------------.((((--((((.....))))............(((.........)))))))((((.....(((((.....)))))...))))................------ ( -20.80, z-score = -1.80, R) >droSec1.super_15 741789 114 + 1954846 CGUUCGGCGGAUGUAGUGUUCUACUCGCUGUCCGAGAUCGUUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUCGCU------ .((((.((((((((.........((((.....)))).((((..((.(((....))).))..))))))))))))...(((((.....)))))...))))................------ ( -29.40, z-score = -1.28, R) >droSim1.chrX_random 431183 114 + 5698898 CGUUCGGCGGAUGUAGUGUUCUACUCGCUGUCCGAGAUCGUUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUCGCU------ .((((.((((((((.........((((.....)))).((((..((.(((....))).))..))))))))))))...(((((.....)))))...))))................------ ( -29.40, z-score = -1.28, R) >consensus CGUUCGGCGGAUGUAGUGUUCUACUCGUUAACCGAGAUCGUUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUCGCU______ .................((((..((((.....))))............(((.........)))))))((((.....(((((.....)))))...))))...................... (-20.08 = -19.92 + -0.16)


| Location | 10,023,066 – 10,023,180 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.41 |
| Shannon entropy | 0.25463 |
| G+C content | 0.42150 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10023066 114 - 22422827 ------AGCGAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAACGAUCUCGGGAAACGAGUAGAACACUACAUCCGCCGAACG ------.(((...(((......((((..((((((.....))))))....))))(((((((.........)))............((((.....))))..))))....))).)))...... ( -28.10, z-score = -1.35, R) >droEre2.scaffold_4690 13656140 104 + 18748788 AGCGAAAAUGAAAAUGAAAAUGGUCCCUGGCCAAGUUAAUUGGCUCUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAACGAUCUCGGUUAACGAGAAAAACG---------------- .(((((.((....)).......(((((.((((((.....))))))..).)))).)))))........................(((((.....)))))......---------------- ( -26.80, z-score = -2.65, R) >droYak2.chrX 18604177 96 - 21770863 ------AGCGAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAACGAUCUCGGUUAACGAG--GAACG---------------- ------.(((((.((....)).((((..((((((.....))))))....)))).)))))........................(((((.....))))--)....---------------- ( -25.50, z-score = -2.07, R) >droSec1.super_15 741789 114 - 1954846 ------AGCGAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAACGAUCUCGGACAGCGAGUAGAACACUACAUCCGCCGAACG ------.(((((.((....)).((((..((((((.....))))))....)))).)))))..........................((((...(.((((((....)))).))).))))... ( -29.00, z-score = -1.45, R) >droSim1.chrX_random 431183 114 - 5698898 ------AGCGAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAACGAUCUCGGACAGCGAGUAGAACACUACAUCCGCCGAACG ------.(((((.((....)).((((..((((((.....))))))....)))).)))))..........................((((...(.((((((....)))).))).))))... ( -29.00, z-score = -1.45, R) >consensus ______AGCGAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAACGAUCUCGGACAACGAGUAGAACACUACAUCCGCCGAACG ......................((((..((((((.....))))))....))))(((((((.........)))............((((.....))))..))))................. (-22.74 = -22.94 + 0.20)

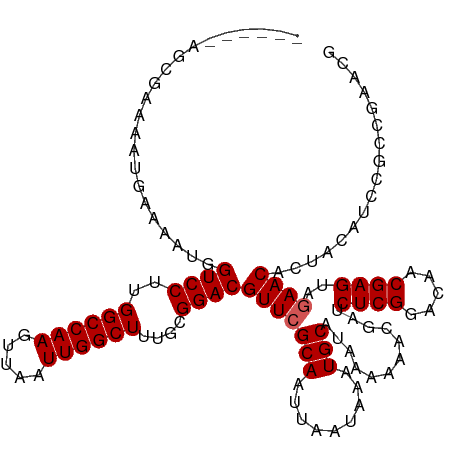
| Location | 10,023,106 – 10,023,198 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 96.61 |
| Shannon entropy | 0.05893 |
| G+C content | 0.41411 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10023106 92 + 22422827 UUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUC------GCUGCAUACUCUCGGGCAGCU .......((((.........))))...((((.....(((((.....)))))...)))).............------(((((...(....).))))). ( -21.20, z-score = -1.40, R) >droEre2.scaffold_4690 13656164 98 - 18748788 UUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAGAGCCAAUUAACUUGGCCAGGGACCAUUUUCAUUUUCAUUUUCGCUGCAUACUCUCGGGCAGCU .......((((.........))))...((((.(.(.(((((.....)))))).)))))...................(((((...(....).))))). ( -21.30, z-score = -0.89, R) >droYak2.chrX 18604199 92 + 21770863 UUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUC------GCUGCAUACUCUCGGGCAGCU .......((((.........))))...((((.....(((((.....)))))...)))).............------(((((...(....).))))). ( -21.20, z-score = -1.40, R) >droSec1.super_15 741829 92 + 1954846 UUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUC------GCUGCAUACUCUCGGGCAGCU .......((((.........))))...((((.....(((((.....)))))...)))).............------(((((...(....).))))). ( -21.20, z-score = -1.40, R) >droSim1.chrX_random 431223 92 + 5698898 UUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUC------GCUGCAUACUCUCGGGCAGCU .......((((.........))))...((((.....(((((.....)))))...)))).............------(((((...(....).))))). ( -21.20, z-score = -1.40, R) >consensus UUUUUUAUGCAUUUAUUAAUUGCGAACGUCCGCAAAGCCAAUUAACUUGGCCAAGGACCAUUUUCAUUUUC______GCUGCAUACUCUCGGGCAGCU .......((((.........))))...((((.....(((((.....)))))...))))...................(((((...(....).))))). (-20.74 = -20.74 + 0.00)

| Location | 10,023,106 – 10,023,198 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 96.61 |
| Shannon entropy | 0.05893 |
| G+C content | 0.41411 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
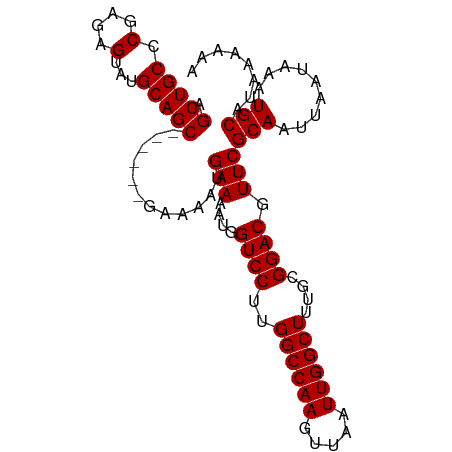
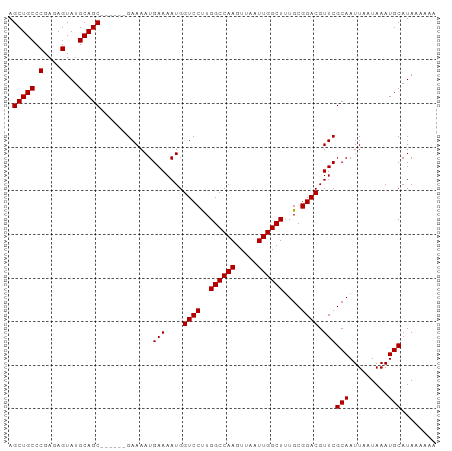
>dm3.chrX 10023106 92 - 22422827 AGCUGCCCGAGAGUAUGCAGC------GAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAA .(((((.(....)...)))))------......(((....((((..((((((.....))))))....)))).)))(((.........)))........ ( -24.50, z-score = -1.26, R) >droEre2.scaffold_4690 13656164 98 + 18748788 AGCUGCCCGAGAGUAUGCAGCGAAAAUGAAAAUGAAAAUGGUCCCUGGCCAAGUUAAUUGGCUCUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAA .(((((.(....)...)))))............(((....(((((.((((((.....))))))..).)))).)))(((.........)))........ ( -24.90, z-score = -1.40, R) >droYak2.chrX 18604199 92 - 21770863 AGCUGCCCGAGAGUAUGCAGC------GAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAA .(((((.(....)...)))))------......(((....((((..((((((.....))))))....)))).)))(((.........)))........ ( -24.50, z-score = -1.26, R) >droSec1.super_15 741829 92 - 1954846 AGCUGCCCGAGAGUAUGCAGC------GAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAA .(((((.(....)...)))))------......(((....((((..((((((.....))))))....)))).)))(((.........)))........ ( -24.50, z-score = -1.26, R) >droSim1.chrX_random 431223 92 - 5698898 AGCUGCCCGAGAGUAUGCAGC------GAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAA .(((((.(....)...)))))------......(((....((((..((((((.....))))))....)))).)))(((.........)))........ ( -24.50, z-score = -1.26, R) >consensus AGCUGCCCGAGAGUAUGCAGC______GAAAAUGAAAAUGGUCCUUGGCCAAGUUAAUUGGCUUUGCGGACGUUCGCAAUUAAUAAAUGCAUAAAAAA .(((((.(....)...)))))............(((....((((..((((((.....))))))....)))).)))(((.........)))........ (-23.00 = -23.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:21 2011