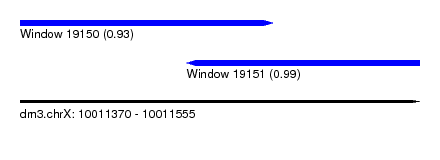
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,011,370 – 10,011,555 |
| Length | 185 |
| Max. P | 0.994792 |

| Location | 10,011,370 – 10,011,487 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Shannon entropy | 0.29656 |
| G+C content | 0.47664 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -24.21 |
| Energy contribution | -23.89 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.08 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10011370 117 + 22422827 GGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGAC---UCAGUCUUGGCUUUUAUUCCCCUCAUUCCCCUUUUUACCACCCAAUGCCACCAUUUGUUUUGUUGGCCAU (((((((..(((((.((((........))))...)))...(((((..---.))))).((((...................................))))......))...))))))).. ( -23.55, z-score = -0.88, R) >droSim1.chrX 7951388 114 + 17042790 GGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGAC---UCAGUCUUGGCUUUUAUCCCCUCUA--CCCCCUUUUACC-CCUCUUGCCACCUUUUGUUUUGUUGGCCAU (((((((..((.(.((((((........(((...)))(..(((((..---.)))))..)................--............-...)))))).).....))...))))))).. ( -24.30, z-score = -1.78, R) >droSec1.super_15 725798 114 + 1954846 GGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGAC---UCAGUCUUGGCUUUCAUCCCCCUUA---UCCCCUUUUACCCCUCUUGCCACCUUUUGUUUUGUUGGCCAU (((((((..((.(.((((((........(((...)))(..(((((..---.)))))..)................---...............)))))).).....))...))))))).. ( -24.30, z-score = -1.62, R) >droYak2.chrX 18593670 108 + 21770863 GGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGACUCAGUAGUCUUGGCUUUUAUCCCC------------UUUUACCCCCCUUGCCCCCUUUUGUUUUGUUGGCCAU (((((((..((.(.((((((........(((...)))(..(((((......)))))..)............------------..........)))))).).....))...))))))).. ( -23.10, z-score = -0.83, R) >droEre2.scaffold_4690 13645310 94 - 18748788 GGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGAC---GCGGUCUUGGCUUUCAUCCCC------------UU-----------GCCCCCUUUUGUUGUGUUGGCCAU (((((((..((((((...(((((.....(((...)))..)))))..)---)))))...(((..........------------..-----------)))............))))))).. ( -24.60, z-score = -0.29, R) >consensus GGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGAC___UCAGUCUUGGCUUUUAUCCCC___A____CC__UUUAAC_CCCCUUGCCACCUUUUGUUUUGUUGGCCAU (((((((..(((((.((((........))))...)))...(((((......)))))..(((...................................))).......))...))))))).. (-24.21 = -23.89 + -0.32)

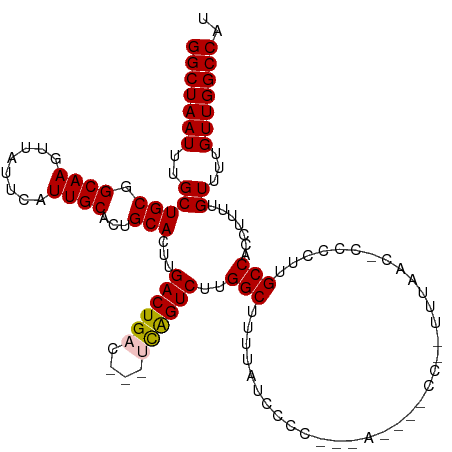
| Location | 10,011,447 – 10,011,555 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Shannon entropy | 0.31706 |
| G+C content | 0.46405 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -34.18 |
| Energy contribution | -35.34 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10011447 108 - 22422827 ------------CCGCUUUUUCUUUAUUGUUUUUGUUCUUUUCUGUUGCUGUCUCAUGUGGCAGAGAGACGUGACAGGAAAUGGCCAACAAAACAAAUGGUGGCAUUGGGUGGUAAAAAG ------------((((((..(((...(((((((((..(((((((((..(.(((((..........))))))..)))))))).)..))..))))))).....)).)..))))))....... ( -29.90, z-score = -1.29, R) >droSim1.chrX 7951463 119 - 17042790 CUGCUUCGGCUGCCGCUUUUUCUUUAUUGUUUUUGUUCUUUUCUGUUGCUGCCUCAUGUGGCGCAGAGACGUGACAGGAAAUGGCCAACAAAACAAAAGGUGGCA-AGAGGGGUAAAAGG .((((((...(((((((((((.(((..((((...((..((((((((..(.(.(((.((.....))))).))..))))))))..)).))))))).)))))))))))-...))))))..... ( -39.00, z-score = -1.45, R) >droSec1.super_15 725873 119 - 1954846 CUGCUUCGGCUGCCGCUUUUUCUUUAUUGUUUUUGUUCUUUUCUGUUGCUUCCUCAUGUGGCGCAGAGACGUGACAGGAAAUGGCCAACAAAACAAAAGGUGGCA-AGAGGGGUAAAAGG .((((((...(((((((((((.(((..((((...((..((((((((..(...(((.((.....)))))..)..))))))))..)).))))))).)))))))))))-...))))))..... ( -38.50, z-score = -1.65, R) >droYak2.chrX 18593739 117 - 21770863 CUGCUUUAGUUGCCCCCUUUUCUUGAUUGUUUUUGUUCUUUUCUGUUGCUGUCUCAUGUGGC--AGAGACGUGACAGGAAAUGGCCAACAAAACAAAAGGGGGCAAGGGGGGUAAAAGG- .((((((..((((((((((((.((..(((((...((..((((((((..(.(((((.......--.))))))..))))))))..)).))))))).))))))))))))..)))))).....- ( -48.80, z-score = -4.40, R) >droEre2.scaffold_4690 13645374 108 + 18748788 CUGCCUCGCUUGCCCCUCUUUCUUGAUUGUUUUUGCUCUUUUCUGUUGCUGUCUCAUGUGGC--AGAGACGUGACAGGAAAUGGCCAACACAACAAAAGGGGGCAAGGGG---------- ....(((.((((((((.((((..((.((((.((.(((.((((((((..(.(((((.......--.))))))..)))))))).))).)).)))))))))))))))))))))---------- ( -47.50, z-score = -4.51, R) >consensus CUGCUUCGGCUGCCGCUUUUUCUUUAUUGUUUUUGUUCUUUUCUGUUGCUGUCUCAUGUGGC__AGAGACGUGACAGGAAAUGGCCAACAAAACAAAAGGUGGCA_AGGGGGGUAAAAGG ...((((...(((((((((((.(((..((((...((..((((((((..(.(((((..........))))))..))))))))..)).))))))).)))))))))))..))))......... (-34.18 = -35.34 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:18 2011